New Pesticidal Diterpenoids from Vellozia gigantea (Velloziaceae), an Endemic Neotropical Plant Living in the Endangered Brazilian Biome Rupestrian Grasslands
Abstract
:1. Introduction
2. Results
2.1. Bioassay-Guided Fractionation
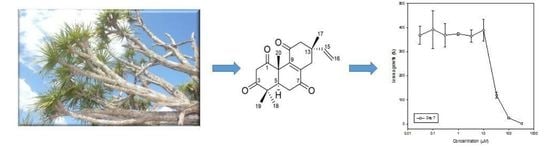
2.2. Identification of Phytotoxic Compounds
2.3. Phytotoxic Activity of the Pure Compounds
2.4. Larval Bioassays
3. Materials and Methods
3.1. Study Area
3.2. Adventitious Roots Extract Production
3.3. Adventitious Roots Extract Fractionation
3.4. NMR Spectroscopy
3.5. HPLC Analysis
3.6. High-Resolution LC-MS Analysis
3.7. Bioassays with Lettuce and Bentgrass
3.8. Bioassay with Lemna Paucicostata (Duckweed)
3.9. Mosquito Larvae Bioassays
4. Discussion
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Lousada, J.M.; Borba, E.L.; Ribeiro, K.T.; Ribeiro, L.C.; Lovato, M.B. Genetic structure and variability of the endemic and vulnerable Vellozia gigantea (Velloziaceae) associated with the landscape in the Espinhaço Range, in southeastern Brazil: implications for conservation. Genetica 2011, 139, 431–440. [Google Scholar] [CrossRef] [PubMed]
- Giulietti, A.M.; Pirani, J.R. Patterns of geographic distribution of some plant species from the Espinhaco Range, Minas Gerais and Bahia, Brazil. In Proceedings of a Workshop on Neotropical Distribution Patterns; Vanzolini, P.E., Heyer, W.R., Eds.; Academia Brasileira de Ciências: Rio de Janeiro, Brazil, 1988; pp. 39–69. [Google Scholar]
- Joly, A.B. Conheça a Vegetação Brasileira; University of São Paulo (USP): São Paulo, Brazil, 1970. [Google Scholar]
- United Nations Educational, Scientific and Cultural Organization (UNESCO). MAB Biosphere Reserves Directory. Available online: http://www.brasilia.unesco.org/noticias/releases/2005/biosferaespinhaco (accessed on 29 September 2014).
- Silveira, F.A.O.; Negreiros, D.; Barbosa, N.P.U.; Buisson, E.; Carmo, F.F.; Carstensen, D.W.; Conceição, A.A.; Cornelissen, T.G.; Ecthernacht, L.; Fernandes, G.W.; et al. Ecology and evolution of plant diversity in the endangered campo rupestre: A neglected conservation priority. Plant Soil 2016, 403, 129–152. [Google Scholar] [CrossRef]
- Mello-Silva, R.; Menezes, N.L. Two new Brazilian Velloziaceae, Vellozia auriculata and Vellozia gigantea, and a key to the related dracenoid species of Vellozia. Novon 1999, 9, 536–541. [Google Scholar] [CrossRef]
- Biodiversitas. Lista da Flora Brasileira Ameaçada de Extinção; Fundação Biodiversitas: Belo Horizonte, Brazil, 2005. [Google Scholar]
- Duke, S.O.; Owens, D.K.; Dayan, F.E. The growing need for biochemical bioherbicides. Am. Chem. Soc. Symp. Ser. 2014, 1172, 31–43. [Google Scholar]
- Pinto, A.C.; Valente, L.M.M.; da Silva, R.S. Norditerpenoids from Vellozia pusilla. Phytochemistry 1988, 27, 3913–3915. [Google Scholar] [CrossRef]
- Pinto, A.C.; Pereira, A.L.; Antunes, O.A.C. RMN 13C de diterpenoids com esqueleto cleistantano. Quim. Nova 1985, 8, 7–13. [Google Scholar]
- Cheng, L.; Ji, K.; Liao, S.; Gan, L.; Yang, L.; Cao, D.; Liu, Y.; Guo, J.; Zhang, P.; Lu, C.; et al. Diterpenoids and Phenanthrenones from the Leaves and Stems of Strophioblachia Fimbricalyx. Tetrahedron Lett. 2016, 21, 2262–2265. [Google Scholar] [CrossRef]
- Michel, A.; Johnson, R.D.; Duke, S.O.; Scheffler, B.E. Dose-response relationships between herbicides with different modes of action and growth of Lemna paucicostata: An improved exotoxicological method. Environ. Toxicol. Chem. 2004, 232, 1074–1079. [Google Scholar] [CrossRef]
- Herbarium of Institute of Biological Science (BHCB) of the Federal University of Minas Gerais, Brazil. Available online: http://sciweb.nybg.org/science2/IndexHerbariorum.asp (accessed on 5 May 2013).
- Dayan, F.E.; Romagni, J.G.; Duke, S.O. Investigating the mode of action of natural phytotoxins. J. Chem. Ecol. 2000, 26, 2079–2094. [Google Scholar] [CrossRef]
- R-Development-Core-Team. R: A Language and Environment for Statistical Computing; 3.2.4 Version; R Foundation for Statistical Computing: Vienna, Austria, 2015. [Google Scholar]
- Ali, A.; Tabanca, N.; Demirci, B.; Baser, K.H.C.; Ellis, J.; Gray, S.; Lackey, B.R.; Murphy, C.; Khan, I.A.; Wedge, D.E. Composition, mosquito larvicidal, biting deterrent and antifungal activity of essential oils of different plant parts of Cupressus arizonica var. glabra (Sudw.) Little (‘Carolina Sapphire’). Nat. Prod. Commun. 2013, 8, 257–260. [Google Scholar] [PubMed]
- Duke, S.O.; Romagni, J.G.; Dayan, F.E. Natural products as sources for new mechanisms of herbicidal action. Crop Protect. 2000, 19, 583–589. [Google Scholar] [CrossRef]
- Tellez, M.R.; Duke, S.O.; Schrader, K.K.; Dayan, F.E.; Romagni, J.G.; Kobaisy, M. Terpenoid-based defense in plants and other organisms. In Lipid Biotechnology; Kuo, T.M., Gardner, H.W., Eds.; Marcel Dekker: New York, NY, USA, 2002; pp. 319–355. [Google Scholar]
- Duke, S.O.; Oliva, A. Mode of action of phytotoxic terpenoids. In Allelopathy: Chemistry and Mode of Action of Allelochemicals; Macías, F.A., Galindo, J.C.G., Molinillo, J.M.G., Eds.; CRC Press: Boca Raton, FL, USA, 2004; pp. 201–216. [Google Scholar]
- Pinto, A.C.; Queiroz, P.P.S.; Garcez, W.S. Diterpenes from Vellozia bicolor. J. Brm. Chem. Soc. 1991, 2, 25–30. [Google Scholar]
- Pinto, A.C.; Rezende, C.M.; Antunes, O.A.; Correia, C.R.D. Three isomeric diterpenes from Vellozia flavicans. Phytochemistry 1996, 42, 767–769. [Google Scholar] [CrossRef]
- Pinto, A.C.; Borges, C. Six diterpenes from Vellozia compacta. Phytochemistry 1983, 22, 2011–2015. [Google Scholar] [CrossRef]
- Morales, M.; Garcia, Q.S.; Siqueira-Silva, A.I.; Silva, M.C.; Munne-Bosch, S. Tocotrienols in Vellozia gigantea leaves: Occurrence and modulation by seasonal and plant size effects. Planta 2014, 240, 437–446. [Google Scholar] [CrossRef] [PubMed]
- Sample Availability: Samples of compound 1, 2, 3, and 4 are available from the authors.



| Compounds | Tested Concentration (µg·mL−1) | Phytotoxicity a | |
|---|---|---|---|
| Lactuca sativa | Agrostis stolonifera | ||
| 1 | 1 | 0 | 0 |
| 10 | 0 | 0 | |
| 100 | 1 | 0 | |
| 1000 | 2 | 3 | |
| 2 | 1 | 0 | 0 |
| 10 | 0 | 0 | |
| 100 | 2 | 1 | |
| 1000 | 3 | 4 | |
| 3 | 1 | 0 | 0 |
| 10 | 0 | 0 | |
| 100 | 0 | 0 | |
| 1000 | 0 | 3 | |
| 4 | 1 | 0 | 0 |
| 10 | 0 | 0 | |
| 100 | 0 | 0 | |
| 1000 | 2 | 3 | |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ferreira, M.C.; Cantrell, C.L.; Duke, S.O.; Ali, A.; Rosa, L.H. New Pesticidal Diterpenoids from Vellozia gigantea (Velloziaceae), an Endemic Neotropical Plant Living in the Endangered Brazilian Biome Rupestrian Grasslands. Molecules 2017, 22, 175. https://doi.org/10.3390/molecules22010175
Ferreira MC, Cantrell CL, Duke SO, Ali A, Rosa LH. New Pesticidal Diterpenoids from Vellozia gigantea (Velloziaceae), an Endemic Neotropical Plant Living in the Endangered Brazilian Biome Rupestrian Grasslands. Molecules. 2017; 22(1):175. https://doi.org/10.3390/molecules22010175
Chicago/Turabian StyleFerreira, Mariana C., Charles L. Cantrell, Stephen O. Duke, Abbas Ali, and Luiz H. Rosa. 2017. "New Pesticidal Diterpenoids from Vellozia gigantea (Velloziaceae), an Endemic Neotropical Plant Living in the Endangered Brazilian Biome Rupestrian Grasslands" Molecules 22, no. 1: 175. https://doi.org/10.3390/molecules22010175
APA StyleFerreira, M. C., Cantrell, C. L., Duke, S. O., Ali, A., & Rosa, L. H. (2017). New Pesticidal Diterpenoids from Vellozia gigantea (Velloziaceae), an Endemic Neotropical Plant Living in the Endangered Brazilian Biome Rupestrian Grasslands. Molecules, 22(1), 175. https://doi.org/10.3390/molecules22010175