A Comparative Genomic and Transcriptomic Survey Provides Novel Insights into N-Acetylserotonin Methyltransferase (ASMT) in Fish
Abstract
:1. Introduction
2. Results
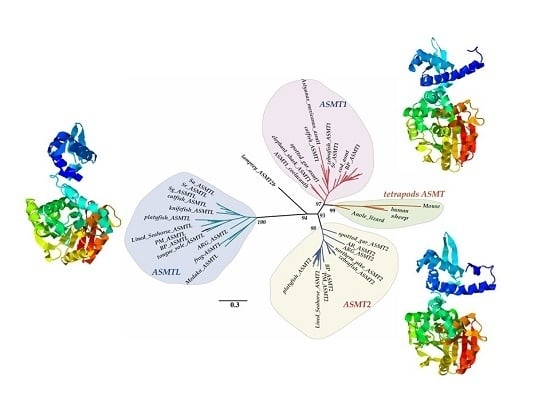
2.1. Copy Number Variation and Phylogenetic Relationships
2.2. Synteny Data
2.3. Structural Analysis of ASMT1, ASMT2 and ASMTL
2.4. The Structure of ASMT Proteins
2.5. Transcription and Cloning of ASMTs
2.6. Predicted Three-Dimensional (3D) Structures of Fish ASMTs
3. Discussion
4. Materials and Methods
4.1. Acquisition of ASMT and ASMTL for Nucleotide and Protein Sequences
4.2. Sequence Alignment and Phylogenetic Analysis
4.3. Analyses of Conserved Synteny and Gene Structures
4.4. Molecular Cloning of Mudskipper ASMT1, ASMT2 and ASMTL Transcripts
4.5. Acquisition of Transcriptomic Data and Quantification of ASMT Transcripts
4.6. Tertiary Structure and Functional Prediction of Each ASMT Protein
5. Conclusions
Supplementary Materials
Acknowledgements
Author Contributions
Conflicts of Interest
References
- Falcón, J.; Besseau, L.; Sauzet, S. Melatonin effects on the hypothalamopituitary axis in fish. Trends Endocrinol. Metab. 2007, 18, 81–88. [Google Scholar] [CrossRef] [PubMed]
- Klein, D.C. Arylalkylamine N-acetyltransferase: “the Timezyme”. J. Biol. Chem. 2007, 282, 4233–4237. [Google Scholar] [CrossRef] [PubMed]
- Klein, D.C.; Coon, S.L.; Roseboom, P.H. The melatonin rhythm-generating enzyme: Molecular regulation of serotonin N-acetyltransferase in the pineal gland. Recent Prog. Horm. Res. 1997, 52, 307–358. [Google Scholar] [PubMed]
- Falcon, J. Cellular circadian clocks in the pineal. Prog. Neurobiol. 1999, 58, 121–162. [Google Scholar] [CrossRef]
- Falcon, J.; Besseau, L.; Boeuf, G. Molecular and cellular regulation of pineal organ responses. Fish Physiol. 2006, 25, 243–306. [Google Scholar]
- Liu, T.; Borjigin, J. N-acetyltransferase is not the rate-limiting enzyme of melatonin synthesis at night. J. Pineal Res. 2005, 39, 91–96. [Google Scholar] [CrossRef] [PubMed]
- Reiter, R.J.; Tan, D.; Terron, M.P. Melatonin and its metabolites: New findings regarding their production and their radical scavenging actions. Acta Biochim. Pol. 2007, 54, 1. [Google Scholar] [PubMed]
- Park, S.; Byeon, Y.; Back, K. Functional analyses of three ASMT gene family members in rice plants. J. Pineal Res. 2013, 55, 409–415. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Li, J.; Ruan, Y. Sequencing ASMT identifies rare mutations in Chinese Han patients with autism. PLoS ONE 2013, 8, e53727. [Google Scholar] [CrossRef] [PubMed]
- Paulin, C.H.; Cazaméa-Catalan, D.; Zilberman-Peled, B. Subfunctionalization of arylalkylamine N-acetyltransferases in the sea bass Dicentrarchus labrax: Two-ones for one two. J. Pineal Res. 2015, 59, 354–364. [Google Scholar] [CrossRef] [PubMed]
- Kang, K.; Kong, K.; Park, S. Molecular cloning of a plant N-acetylserotonin methyltransferase and its expression characteristics in rice. J. Pineal Res. 2011, 50, 304–309. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Byeon, Y.; Kim, Y.S. Kinetic analysis of purified recombinant rice N-acetylserotonin methyltransferase and peak melatonin production in etiolated rice shoots. J. Pineal Res. 2013, 54, 139–144. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, I.R.; Mazuruk, K.; Schoen, T.J. Structural analysis of the human hydroxyindole- O-methyltransferase gene. Presence of two distinct promoters. J. Biol. Chem. 1994, 269, 31969–31977. [Google Scholar] [PubMed]
- Botros, H.G.; Legrand, P.; Pagan, C. Crystal structure and functional mapping of human ASMT, the last enzyme of the melatonin synthesis pathway. J. Pineal Res. 2013, 54, 46–57. [Google Scholar] [CrossRef] [PubMed]
- Velarde, E.; Cerdá-Reverter, J.M.; Alonso-Gómez, A.L. Melatonin-synthesizing enzymes in pineal, retina, liver, and gut of the goldfish (Carassius): mRNA expression pattern and regulation of daily rhythms by lighting conditions. Chronobiol. Int. 2010, 27, 1178–1201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, Z.A.; Yumnamcha, T.; Rajiv, C. Melatonin biosynthesizing enzyme genes and clock genes in ovary and whole brain of zebrafish (Danio rerio): Differential expression and a possible interplay. Gen. Comp. Endocrinol. 2016, 233, 16–31. [Google Scholar] [CrossRef] [PubMed]
- Muñoz-Pérez, J.L.; López-Patiño, M.A.; Álvarez-Otero, R. Characterization of melatonin synthesis in the gastrointestinal tract of rainbow trout (Oncorhynchus mykiss): Distribution, relation with serotonin, daily rhythms and photoperiod regulation. J. Comp. Physiol. B 2016, 186, 471–484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanjita Devi, H.; Rajiv, C.; Mondal, G. Melatonin bio-synthesizing enzyme genes (Tph1, Aanat1, Aanat2, and Hiomt) and their temporal pattern of expression in brain and gut of a tropical carp in natural environmental conditions. Cogent Biol. 2016, 2, 1230337. [Google Scholar] [CrossRef]
- Mukherjee, S.; Maitra, S.K. Gut melatonin in vertebrates: Chronobiology and physiology. Front. Endocrinol. 2015, 6, 112. [Google Scholar] [CrossRef] [PubMed]
- Velarde, E.; Delgado, M.J.; Alonso-Gómez, A.L. Serotonin-induced contraction in isolated intestine from a teleost fish (Carassius auratus): Characterization and interactions with melatonin. Neurogastroenterol. Motil. 2010, 22, e364–e373. [Google Scholar] [CrossRef] [PubMed]
- Velarde, E.; Haque, R.; Iuvone, P.M. Circadian clock genes of goldfish, Carassius auratus: cDNA cloning and rhythmic expression of period and cryptochrome transcripts in retina, liver, and gut. J. Biol. Rhythm. 2009, 24, 104–113. [Google Scholar] [CrossRef] [PubMed]
- Ried, K.; Rao, E.; Schiebel, K. Gene duplications as a recurrent theme in the evolution of the human pseudoautosomal region 1: Isolation of the gene ASMTL. Hum. Mol. Genet. 1998, 7, 1771–1778. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Huang, Y.; Li, X. Fish-T1K (Transcriptomes of 1,000 Fishes) Project: Large-scale transcriptome data for fish evolution studies. GigaScience 2016, 5, 18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.; Chen, X.; Bai, J. The Sinocyclocheilus cavefish genome provides insights into cave adaptation. BMC Biol. 2016, 14, 1. [Google Scholar] [CrossRef] [PubMed]
- Zubieta, C.; He, X.Z.; Dixon, R.A. Structures of two natural product methyltransferases reveal the basis for substrate specificity in plant O-methyltransferases. Nat. Struct. Mol. Biol. 2001, 8, 271–279. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.J.; Deavours, B.E.; Richard, S.B. Structural basis for dual functionality of isoflavonoid O-methyltransferases in the evolution of plant defense responses. Plant Cell 2006, 18, 3656–3669. [Google Scholar] [CrossRef] [PubMed]
- Jaillon, O.; Aury, J.M.; Brunet, F. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature 2004, 431, 946–957. [Google Scholar] [CrossRef] [PubMed]
- Kasahara, M.; Naruse, K.; Sasaki, S. The medaka draft genome and insights into vertebrate genome evolution. Nature 2007, 447, 714–719. [Google Scholar] [CrossRef] [PubMed]
- Nakatani, Y.; Takeda, H.; Kohara, Y. Reconstruction of the vertebrate ancestral genome reveals dynamic genome reorganization in early vertebrates. Genome Res. 2007, 17, 1254–1265. [Google Scholar] [CrossRef] [PubMed]
- Mayfield-Jones, D.; Washburn, J.D.; Arias, T. Watching the grin fade: Tracing the effects of polyploidy on different evolutionary time scales. Semin. Cell Dev. Biol. 2013, 24, 320–331. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; You, X.; Bian, C. Molecular evolution of aralkylamine N-acetyltransferase in fish: A genomic survey. Int. J. Mol. Sci. 2016, 17, 51. [Google Scholar] [CrossRef] [PubMed]
- Davidson, W.S.; Koop, B.F.; Jones, S.J.M. Sequencing the genome of the Atlantic salmon (Salmo salar). Genome Biol. 2010, 11, 403. [Google Scholar] [PubMed]
- Zilberman-Peled, B.; Bransburg-Zabary, S.; Klein, D.C. Molecular evolution of multiple arylalkylamine N-acetyltransferase (AANAT) in fish. Mar. Drugs 2011, 9, 906–921. [Google Scholar] [CrossRef] [PubMed]
- Force, A.; Lynch, M.; Pickett, F.B. Preservation of duplicate genes by complementary, degenerative mutations. Genetics 1999, 151, 1531–1545. [Google Scholar] [PubMed]
- Lien, S.; Koop, B.F.; Sandve, S.R. The Atlantic salmon genome provides insights into rediploidization. Nature 2016, 533, 200–205. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, I.; Maitra, S.K.; Verster, G.C. Melatonin in the Promotion of Health. In Melatonin Time Line: From Discovery to Therapy; Taylor and Francis: Boca Raton, FL, USA, 2012; pp. 1–60. [Google Scholar]
- Jimenez-Jorge, S.; Guerrero, J.M.; Jimenez-Caliani, A.J. Evidence for melatonin synthesis in the rat brain during development. J. Pineal Res. 2007, 42, 240–246. [Google Scholar] [CrossRef] [PubMed]
- Skolnick, J.; Fetrow, J.S. From genes to protein structure and function: Novel applications of computational approaches in the genomic era. Trends Biotechnol. 2000, 18, 34–39. [Google Scholar] [CrossRef]
- Pevsner, J. Basic Local Alignment Search Tool (BLAST). In Bioinformatics and Functional Genomics, 2nd ed.; Wiley: Hoboken, NJ, USA, 2009; pp. 100–138. [Google Scholar]
- Birney, E.; Clamp, M.; Durbin, R. GeneWise and Genomewise. Genome Res. 2004, 14, 988–995. [Google Scholar] [CrossRef] [PubMed]
- Yamada, K.D.; Tomii, K.; Katoh, K. Application of the MAFFT sequence alignment program to large data-reexamination of the usefulness of chained guide trees. Bioinformatics 2016, 32, 3246–3251. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 2006, 22, 2688–2690. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A.; Hoover, P.; Rougemont, J. A rapid bootstrap algorithm for the RAxML Web servers. Syst. Biol. 2008, 57, 758–771. [Google Scholar] [CrossRef] [PubMed]
- Price, M.N.; Dehal, P.; Arkin, A.P. FastTree: Computing Large Minimum-Evolution Trees with Profiles instead of a Distance Matrix. Mol. Biol. Evol. 2009, 26, 1641–1650. [Google Scholar] [CrossRef] [PubMed]
- Jones, F.C.; Grabherr, M.G.; Chan, Y.F. The genomic basis of adaptive evolution in three spine sticklebacks. Nature 2012, 484, 55–61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- You, X.; Bian, C.; Zan, Q. Mudskipper genomes provide insights into the terrestrial adaptation of amphibious fishes. Nat. Commun. 2014, 5, 5594. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Williams, B.A.; Pertea, G. Transcript assembly and abundance estimation from RNA-Seq reveals thousands of new transcripts and switching among isoforms. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Yan, R.; Roy, A. The I-TASSER Suite: Protein structure and function prediction. Nat. Methods 2015, 12, 7–8. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are available from the authors. |







| Common Name | Species Name | ASMT1 | ASMT2 | ASMTL |
|---|---|---|---|---|
| Golden-line fishes (Sa) 1 | Sinocyclocheilus anshuiensis | 1 | 2 | 1 |
| (Sg) 1 | Sinocyclocheilus grahami | 1 | 2 | 1 |
| (Sr) 1 | Sinocyclocheilus rhinocerous | 1 | 2 | 1 |
| Large yellow croaker | Larimichthys crocea | 1 | 1 | 1 |
| Northern snakehead | Channa argus | 1 | 1 | 1 |
| Giant-fin mudskipper | Periophthalmus magnuspinnatus | 1 | 1 | 1 |
| Blue-spotted mudskipper | Boleophthalmus pectinirosris | 1 | 1 | 1 |
| Lined seahorse | Hippocampus erectus | 1 | 1 | 1 |
| Kanglang fish | Anabarilius grahami | 1 | 1 | 1 |
| Black porgy | Acanthopagrus schlegelii | 1 | 1 | 1 |
| Southern platyfish | Xiphophorus maculatus | 1 | 1 | 1 |
| Bicolor damselfish | Stegastes partitus | 1 | 1 | 1 |
| Red-bellied piranha | Pygocentrus nattereri | 1 | 1 | 1 |
| Channel catfish | Ictalurus punetaus | 1 | 1 | 1 |
| Green arowana | Scleropages formosus | 1 | 1 | 1 |
| Golden arowana | Scleropages formosus | 1 | 1 | - |
| Red arowana | Scleropages formosus | 1 | 1 | - |
| Longjaw grenadier anchovy | Coilia macrognathos | 1 | 1 | - |
| Northern pike | Esox lucius | 1 | 1 | - |
| Atlantic salmon | Salmo salar | 1 | - | - |
| Elephant shark | Callorhinchus milii | 1 | - | - |
| Tissue | Gene | Sg | Sr | Sa |
|---|---|---|---|---|
| Eye | ASMT1 | 30.57 | 13.50 | 1.62 |
| ASMT2a | 18.14 | 3.15 | 0.90 | |
| ASMT2b | 0.00 | 0.00 | 8.57 | |
| ASMTL | 0.00 | 3.35 | 122.11 | |
| Skin | ASMT1 | 0.09 | 0.07 | 0.19 |
| ASMT2a | 7.36 | 2.71 | 2.07 | |
| ASMT2b | 0.00 | 0.35 | 5.02 | |
| ASMTL | 0.00 | 4.55 | 37.71 | |
| Liver | ASMT1 | 0.00 | 0.00 | 0.00 |
| ASMT2a | 1.30 | 0.96 | 1.59 | |
| ASMT2b | 0.00 | 0.11 | 5.46 | |
| ASMTL | 0.00 | 71.22 | 118.35 | |
| Gonad | ASMT1 | 0.00 | 0.00 | 0.42 |
| ASMT2a | 0.00 | 0.48 | 0.21 | |
| ASMT2b | 0.00 | 0.05 | 34.99 | |
| ASMTL | 0.00 | 17.71 | 14.78 |
| Tissue | Gene | PM | BP |
|---|---|---|---|
| Brain | ASMT1 | 1.31 | 0.98 |
| ASMT2 | 8.47 | 2.43 | |
| ASMTL | 2.05 | 4.51 | |
| Gill | ASMT1 | 0.00 | 0.00 |
| ASMT2 | 2.43 | 2.43 | |
| ASMTL | 2.76 | 1.54 | |
| Liver | ASMT1 | 0.00 | 0.37 |
| ASMT2 | 3.09 | 1.73 | |
| ASMTL | 7.15 | 1.63 | |
| Muscle | ASMT1 | 0.00 | 0.00 |
| ASMT2 | 5.72 | 1.07 | |
| ASMTL | 3.01 | 1.64 | |
| Skin | ASMT1 | 0.00 | 0.00 |
| ASMT2 | 6.47 | 2.37 | |
| ASMTL | 4.93 | 0.85 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, K.; Ruan, Z.; Li, J.; Bian, C.; You, X.; Coon, S.L.; Shi, Q. A Comparative Genomic and Transcriptomic Survey Provides Novel Insights into N-Acetylserotonin Methyltransferase (ASMT) in Fish. Molecules 2017, 22, 1653. https://doi.org/10.3390/molecules22101653
Zhang K, Ruan Z, Li J, Bian C, You X, Coon SL, Shi Q. A Comparative Genomic and Transcriptomic Survey Provides Novel Insights into N-Acetylserotonin Methyltransferase (ASMT) in Fish. Molecules. 2017; 22(10):1653. https://doi.org/10.3390/molecules22101653
Chicago/Turabian StyleZhang, Kai, Zhiqiang Ruan, Jia Li, Chao Bian, Xinxin You, Steven L. Coon, and Qiong Shi. 2017. "A Comparative Genomic and Transcriptomic Survey Provides Novel Insights into N-Acetylserotonin Methyltransferase (ASMT) in Fish" Molecules 22, no. 10: 1653. https://doi.org/10.3390/molecules22101653
APA StyleZhang, K., Ruan, Z., Li, J., Bian, C., You, X., Coon, S. L., & Shi, Q. (2017). A Comparative Genomic and Transcriptomic Survey Provides Novel Insights into N-Acetylserotonin Methyltransferase (ASMT) in Fish. Molecules, 22(10), 1653. https://doi.org/10.3390/molecules22101653