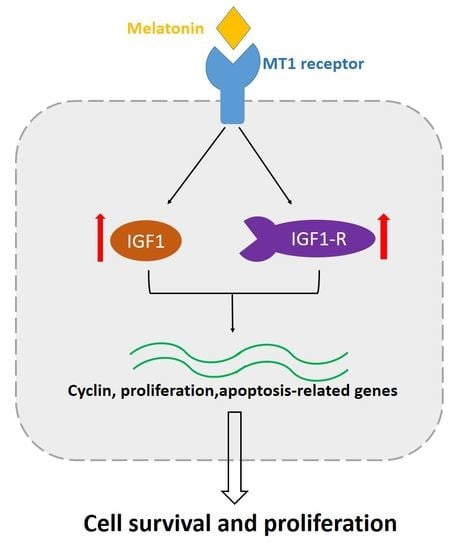
The Regulatory Mechanism of MLT/MT1 Signaling on the Growth of Antler Mesenchymal Cells
Abstract
:1. Introduction
2. Results
2.1. Effect of Melatonin on Cell Proliferation
2.2. The Location and Expression of MT1 in Antler Mesenchymal Cells
2.3. Construction of the Interference Fragment of MT1 Using Plasmid DNA
2.4. Effects of MLT/MT1 Knock-Down on the Viability of Mesenchymal Cells
2.5. Effects of MT1 Knock-Down on Cell Proliferation-Related Gene Expression
2.6. Effects of MT1 Knock-Down on IGF/IGF1-R Signaling
3. Discussion
4. Materials and Methods
4.1. Sample Collection and Culture of Mesenchymal Cells
4.2. RNAi Plasmid Restructuring of MT1
4.3. Western Blot Assay
4.4. Cell Proliferation Test
4.5. Immunofluorescence Assay
4.6. Real-Time PCR Analysis
4.7. Enzyme-Linked Immunosorbent Assay (ELISA) Assay
4.8. Data Analysis and Statistics
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Price, J.; Allen, S. Exploring the mechanisms regulating regeneration of deer antlers. Proc. R. Soc. Lond. B 2004, 359, 809–822. [Google Scholar] [CrossRef] [PubMed]
- Kierdorf, U.; Kierdorf, H.; Schultz, M.; Rolf, H.J. Histological structure of antlers in castrated male fallow deer (Dama dama). Anat. Rec. A 2004, 281A, 1352–1362. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Zhao, H.; Liu, Z.; Mcmahon, C. Deer antler—A novel model for studying organ regeneration in mammals. Int. J. Biochem. Cell Biol. 2014, 56, 111–122. [Google Scholar] [CrossRef] [PubMed]
- Kierdorf, U.; Kierdorf, H.; Szuwart, T. Deer antler regeneration: Cells, concepts, and controversies. J. Morphol. 2007, 268, 726–738. [Google Scholar] [CrossRef] [PubMed]
- Rolf, H.J.; Kierdorf, U.; Kierdorf, H.; Schulz, J.; Seymour, N.; Schliephake, H.; Napp, J.; Niebert, S.; Wölfel, H.; Wiese, K.G. Localization and characterization of STRO-1 cells in the deer pedicle and regenerating antler. PLoS ONE 2008, 3, e2064. [Google Scholar] [CrossRef] [PubMed]
- Molnár, A.; Gyurján, I.; Korpos, E.; Borsy, A.; Stéger, V.; Buzás, Z.; Kiss, I.; Zomborszky, Z.; Papp, P.; Deák, F.; et al. Identification of differentially expressed genes in the developing antler of red deer Cervus elaphus. Mol. Genet. Genom. 2007, 277, 237–248. [Google Scholar] [CrossRef] [PubMed]
- Kierdorf, U.; Kierdorf, H. Deer antlers-a model of mammalian appendage regeneration: An extensive review. Gerontology 2011, 57, 53–65. [Google Scholar] [CrossRef] [PubMed]
- Suttie, J.M.; Gluckman, P.D.; Butler, J.H.; Fennessy, P.F.; Corson, I.D.; Laas, F.J. Insulin-like growth factor 1 (IGF-1) antler-stimulating hormone. Endocrinology 1985, 116, 846–848. [Google Scholar] [CrossRef] [PubMed]
- Price, J.S.; Oyajobi, B.O.; Oreffo, R.O.C.; Russel, R.G. Cells cultured from the growing tip of red deer antler express alkaline phosphatase and proliferate in response to insulin-like growth factor-I. J. Endocrinol. 1994, 143, R9–R16. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Jeon, B.; Kang, S.; Oh, M.; Kim, M.; Jang, S.; Park, P.; Kim, S.; Moon, S. Study on the changes in enzyme and insulin-like growth factor-1 concentrations in blood serum and growth characteristics of velvet antler during the antler growth period in sika deer (Cervus nippon). Asian Austral. J. Anim. 2015, 28, 1303–1308. [Google Scholar] [CrossRef] [PubMed]
- Sadighi, M.; Haines, S.R.; Skottner, A.; Harris, A.J.; Suttie, J.M. Effects of insulin-like growth factor-I (IGF-I) and IGF-II on the growth of antler cells in vitro. J. Endocrinol. 1994, 143, 461–469. [Google Scholar] [CrossRef] [PubMed]
- Gu, L.; Mo, E.; Yang, Z.; Zhu, X.; Fang, Z.; Sun, B.; Wang, C.; Bao, J.; Sung, C. Expression and localization of insulin-like growth factor-I in four parts of the red deer antler. Growth Factors 2007, 25, 264–279. [Google Scholar] [CrossRef] [PubMed]
- Jarzynka, M.J.; Passey, D.K.; Johnson, D.A.; Konduru, N.V.; Fitz, N.F.; Radio, N.M.; Rasenick, M.; Benloucif, S.; Melan, M.A.; Witt-Enderby, P.A. Microtubules modulate melatonin receptors involved in phase-shifting circadian activity rhythms: In vitro and in vivo evidence. J. Pineal Res. 2009, 46, 161–171. [Google Scholar] [CrossRef] [PubMed]
- Cardinali, D.P.; Srinivasan, V.; Brzezinski, A.; Brown, G.M. Melatonin and its analogs in insomnia and depression. J. Pineal Res. 2012, 52, 365–375. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Garcia, A.; Mayo, J.C.; Hevia, D.; Quiros-Gonzalez, I.; Navarro, M.; Sainz, R.M. Phenotypic changes caused by melatonin increased sensitivity of prostate cancer cells to cytokine-induced apoptosis. J. Pineal Res. 2013, 54, 33–45. [Google Scholar] [CrossRef] [PubMed]
- Gurer-Orhan, H.; Suzen, S. Melatonin, its metabolites and its synthetic analogs as multi-faceted compounds: Antioxidant, prooxidant and inhibitor of bioactivation reactions. Curr. Med. Chem. 2015, 22, 490–499. [Google Scholar] [CrossRef] [PubMed]
- Marková, M.; Adámeková, E.; Kubatka, P.; Bojková, B.; Ahlersová, E.; Ahlers, I. Effect of prolonged melatonin application on metabolic parameters and organ weights in young male and female Sprague-Dawley rats. Acta Vet. Brno 2003, 72, 163–173. [Google Scholar] [CrossRef]
- De Berardis, D.; Marini, S.; Fornaro, M.; Srinivasan, V.; Iasevoli, F.; Tomasetti, C.; Valchera, A.; Perna, G.; Quera-Salva, M.A.; Martinotti, G.; et al. The melatonergic system in mood and anxiety disorders and the role of agomelatine: Implications for clinical practice. Int. J. Mol. Sci. 2013, 14, 12458–12483. [Google Scholar] [CrossRef] [PubMed]
- De Berardis, D.; Fornaro, M.; Serroni, N.; Campanella, D.; Rapini, G.; Olivieri, L.; Srinivasan, V.; Iasevoli, F.; Tomasetti, C.; de Bartolomeis, A.; et al. Agomelatine beyond borders: Current evidences of its efficacy in disorders other than major depression. Int. J. Mol. Sci. 2013, 16, 1111–1130. [Google Scholar] [CrossRef] [PubMed]
- Kubatka, P.; Bojková, B.; Mciková-Kalická, K.; Mníchová-Chamilová, M.; Adámeková, E.; Ahlers, I.; Ahlersová, E.; Cermáková, M. Effects of tamoxifen and melatonin on mammary gland cancer induced by N-methyl-N-nitrosourea and by 7, 12-dimethylbenz (a) anthracene, respectively, in female Sprague-Dawley rats. Folia Biol. (Praha) 2001, 47, 5–10. [Google Scholar] [PubMed]
- Mayo, J.C.; Sainz, R.M.; González Menéndez, P.; Cepas, V.; Tan, D.X.; Reiter, R.J. Melatonin and sirtuins: A “not-so unexpected” relationship. J. Pineal Res. 2017, 62. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Wang, J.; Zhang, Z.; Yang, M.; Li, Y.; Tian, X.; Ma, T.; Tao, J.; Zhu, K.; Song, Y.; et al. Mitochondria synthesize melatonin to ameliorate its function and improve mice oocyte’s quality under in vitro conditions. Int. J. Mol. Sci. 2016, 17, 939. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Wang, J.; Li, Y.; Zhu, K.; Xu, Z.; Song, Y.; Song, Y.; Liu, G. Melatonin-related genes expressed in the mouse uterus during early gestation promote embryo implantation. J. Pineal Res. 2015, 58, 300–309. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Ma, T.; Shi, J.; Zhang, Z.; Wang, J.; Zhu, K.; Li, Y.; Yang, M.; Song, Y.; Liu, G. Melatonin and its receptor MT1 are involved in the downstream reaction to luteinizing hormone and participate in the regulation of luteinization in different species. J. Pineal Res. 2016, 61, 279–290. [Google Scholar] [CrossRef] [PubMed]
- De Berardis, D.; Fornaro, M.; Orsolini, L.; Iasevoli, F.; Tomasetti, C.; de Bartolomeis, A.; Serroni, N.; de Lauretis, I.; Girinelli, G.; Mazza, M.; et al. Effect of agomelatine treatment on C-reactive protein levels in patients with major depressive disorder: An exploratory study in “real-world,” everyday clinical practice. CNS Spectr. 2017, 22, 342–347. [Google Scholar] [CrossRef] [PubMed]
- Martinotti, G.; Pettorruso, M.; De Berardis, D.; Varasano, P.A.; Lucidi Pressanti, G.; de Remigis, V.; Valchera, A.; Ricci, V.; di Nicola, M.; Janiri, L.; et al. Agomelatine increases BDNF serum levels in depressed patients in correlation with the improvement of depressive symptoms. Int. J. Neuropsychopharmacol. 2016, 19. [Google Scholar] [CrossRef] [PubMed]
- Wilson, P.R.; Walker, I.H.; Bond, D.B.; Middleberg, A.; Staple, L.D. Field evaluation of melatonin implants to advance the breeding season in l-year-old farmed red deer hinds. N. Z. Vet. J. 1991, 39, 23–28. [Google Scholar] [CrossRef] [PubMed]
- Bubenik, G.A.; Smith, P.S.; Schams, D. The effect of orally administered melatonin on the seasonality of deer pelage exchange, antler development, LH, FSH, prolactin, testosterone, T3, T4, cortisol and alkaline phosphatase. J. Pineal Res. 1986, 3, 331–349. [Google Scholar] [CrossRef] [PubMed]
- Webster, J.R.; Suttie, J.M.; Corson, I.D. Effects of melatonin implants on reproductive seasonality of male red deer (Cervus elaphus). J. Reprod. Fertil. 1991, 92, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Suttie, J.M.; Breier, B.H.; Gluckman, P.D.; Littlejohn, R.P.; Webster, J.R. Effects of melatonin implants on insulin-like growth factor 1 in male red deer (Cervus elaphus). Gen. Comp. Endocrinol. 1992, 87, 111–119. [Google Scholar] [CrossRef]
- Yang, F.F.; Zhang, P.; Huo, L.J.; Riaz, H.; Yang, L.G.; Zhang, S.J.; Xiong, J.J. The association of single nucleotide polymorphism in the IGF1, IGF2 and IGF1R with antler yield in Sika deer. Pak. Vet. J. 2014, 34, 469–473. [Google Scholar]
- Yang, F.F.; Huo, L.J.; Yang, L.G.; Riaz, H.; Xiong, L.R.; Chen, J.G.; Zhang, S.J.; Xiong, J.J. Association between melatonin receptor 1A (MTNR1A) gene single-nucleotide polymorphisms and the velvet antler yield of Sika deer. Mol. Biol. Rep. 2014, 41, 1901–1906. [Google Scholar] [CrossRef] [PubMed]
- Gyurján, I.; Molnár, A.; Borsy, A.; Stéger, V.; Hackler, L.; Zomborszky, Z.; Papp, P.; Duda, E.; Deák, F.; Lakatos, P.; et al. Gene expression dynamics in deer antler: Mesenchymal differentiation toward chondrogenesis. Mol. Genet. Genom. 2007, 277, 221–235. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Yang, F.; Haines, S.; Zhao, H.; Wang, W.; Xing, X.; Sun, H.; Chu, W.; Lu, X.; Liu, L.; et al. Stem cells responsible for deer antler regeneration are unable to recapitulate the process of first antler development—Revealed through intradermal and subcutaneous tissue transplantation. J. Exp. Zool. Part B 2010, 314B, 552–570. [Google Scholar] [CrossRef] [PubMed]
- Cegielski, M.; Izykowska, I.; Podhorska-Okolow, M.; Gworys, B.; Zabel, M.; Dziegiel, P. Histological studies of growing and mature antlers of red deer stags (Cervus elaphus). Anat. Histol. Embryol. 2009, 38, 184–188. [Google Scholar] [CrossRef] [PubMed]
- Pita-Thomas, W.; Barroso-García, G.; Moral, V.; Hackett, A.R.; Cavalli, V.; Nieto-Diaz, M. Identification of axon growth promoters in the secretome of the deer antler velvet. Neuroscience 2016, 340, 333–344. [Google Scholar] [CrossRef] [PubMed]
- Manchester, L.C.; Coto Montes, A.; Boga, J.A.; Andersen, L.P.H.; Zhou, Z.; Galano, A.; Vriend, J.; Tan, D.X.; Reiter, R.J. Melatonin: An ancient molecule that makes oxygen metabolically tolerable. J. Pineal Res. 2015, 59, 403–419. [Google Scholar] [CrossRef] [PubMed]
- Reiter, R.J.; Tan, D.X.; Maldonado, M.D. Melatonin as an antioxidant: Physiology versus pharmacology. J. Pineal Res. 2005, 39, 215–216. [Google Scholar] [CrossRef] [PubMed]
- Emet, M.; Ozcan, H.; Ozel, L.; Yayla, M.; Halici, Z.; Hacimuftuoglu, A. A review of melatonin, its receptors and drugs. Eur. J. Med. 2016, 48, 135–141. [Google Scholar] [CrossRef] [PubMed]
- Reiter, R.J.; Rosales-Corral, S.A.; Manchester, L.C.; Liu, X.; Tan, D.X. Melatonin in the biliary tract and liver: Health implications. Curr. Pharm. Design 2014, 20, 4788–4801. [Google Scholar] [CrossRef]
- Simko, F.; Baka, T.; Paulis, L.; Reiter, R.J. Elevated heart rate and nondipping heart rate as potential targets for melatonin: A review. J. Pineal Res. 2016, 61, 127–137. [Google Scholar] [CrossRef] [PubMed]
- Bubenik, G.A.; Smith, P.S. Circadian and circannual rhythms of melatonin in plasma of male white-tailed deer and the effect of oral administration of melatonin. J. Exp. Zool. Part A 1987, 241, 81–89. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.S.; Yin, T.C.; Sung, P.H.; Chiang, J.Y.; Sun, C.K.; Yip, H.K. Melatonin enhances survival and preserves functional integrity of stem cells: A review. J. Pineal Res. 2017. [Google Scholar] [CrossRef] [PubMed]
- Tanabe, M.; Tamura, H.; Taketani, T.; Okada, M.; Lee, L.; Tamura, I.; Sugino, N. Melatonin protects the integrity of granulosa cells by reducing oxidative stress in nuclei, mitochondria, and plasma membranes in mice. J. Reprod. Dev. 2015, 61, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Song, C.; Peng, W.; Yin, S.; Zhao, J.; Fu, B.; Zhang, J.; Zhang, Y. Melatonin improves age-induced fertility decline and attenuates ovarian mitochondrial oxidative stress in mice. Sci. Rep. 2016, 6, 35165. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.Q.; Zhang, H.L.; Duan, C.; Geng, C.; Wang, S.; Yu, K.; Yue, Z.P.; Guo, B. IGF1 regulates RUNX1 expression via IRS1/2: Implications for antler chondrocyte differentiation. Cell Cycle 2017, 16, 522–532. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Li, T.; Wu, L.; Li, M.; Meng, X. Identification of microRNA-18a as a novel regulator of the insulin-like growth factor-1 in the proliferation and regeneration of deer antler. Biotechnol. Lett. 2014, 36, 703–710. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are not available from the authors. |






| Name | Sense/Antisense | Sequence Sense |
|---|---|---|
| shRNA1 | Sense | 5′-GATCCGGTCAATAGAGAATTAACAGTTCAAGAG ACTGTTAATTCTCTATTGACCTTTTTTAAGCTTG-3′ |
| Antisense | 5′-AATTCAAGCTTAAAAAAGGTCAATAGAGAATTA ACAGTCTCTTGAACTGTTAATTCTCTATTGACCG-3′ | |
| shRNA2 | Sense | 5′-GATCCGCGTAGCAGATAGAATTAAATTCAAGAGA TTTAATTCTATCTGCTACGCTTTTTTAAGCTTG-3′ |
| Antisense | 5′-AATTCAAGCTTAAAAAAGCGTAGCAGATAGAAT TAAATCTCTTGAATTTAATTCTATCTGCTACGCG-3′ | |
| shRNA3 | Sense | 5′-GATCCGGGATCTATTCCTGCACCTTCATTCAAGAGA TGAAGGTGCAGGAATAGATCCCTTTTTTAAGCTTG-3′ |
| Antisense | 5′-AATTCAAGCTTAAAAAAGGGATCTATTCCTGCACC TTCATCTCTTGAATGAAGGTGCAGGAATAGATCCCG-3′ |
| Gene | Primer Sequence (5′-3′) | Tm (°C) | Product Size (bp) |
|---|---|---|---|
| β-actin | F: TGACCCTTAAGTACCCCATCGA | 60 | 85 bp |
| R: TTGTAGAAGGTGTGGTGCCAGAT | |||
| MT1 | F: TGGCTGTTTGTGGCCAGTTA | 60 | 158 bp |
| R: ACGTGATTGGAGCTATCCGC | |||
| IGF1 | F: TGTGATTTCTTGAAGCAGGTGA | 60 | 96 bp |
| R: CGTGGCAGAGCTGGTGAAG | |||
| IGF-1R | F: GCACCATCTTCAAAGGCAATC | 60 | 95 bp |
| R: GAGACCAAGGCGTGGGAGT | |||
| Cyclin D1 | F: GCGCAGACCTTCGTTGCCCT | 60 | 123 bp |
| R: GCCGTTGGCGCTTCCCAGAT | |||
| PCNA | F: CCTTGGTGCAGCTAACCCTT | 60 | 94 bp |
| R: TTGGACATGCTGGTGAGGTT | |||
| P21 | F: GACCACTTGGACCTGTCGCT | 60 | 183 bp |
| R: GGGTTAGGGCTTCCTCTTGG | |||
| P27 | F: AGTGTCTAACGGGAGTCCGA | 60 | 213 bp |
| R: CACTCGTACTTGCCCTCCAG | |||
| P53 | F: GAAGACCTACCCTGGCAATTAC | 60 | 103 bp |
| R: AGAACAGCTTGTTAAGGGAAGG |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, F.; He, C.; Sun, X.; Wang, J.; Luo, C.; Liu, G.; Yang, L.; Xiong, J.; Huo, L. The Regulatory Mechanism of MLT/MT1 Signaling on the Growth of Antler Mesenchymal Cells. Molecules 2017, 22, 1793. https://doi.org/10.3390/molecules22101793
Yang F, He C, Sun X, Wang J, Luo C, Liu G, Yang L, Xiong J, Huo L. The Regulatory Mechanism of MLT/MT1 Signaling on the Growth of Antler Mesenchymal Cells. Molecules. 2017; 22(10):1793. https://doi.org/10.3390/molecules22101793
Chicago/Turabian StyleYang, Feifei, Changjiu He, Xuyang Sun, Jing Wang, Can Luo, Guoshi Liu, Liguo Yang, Jiajun Xiong, and Lijun Huo. 2017. "The Regulatory Mechanism of MLT/MT1 Signaling on the Growth of Antler Mesenchymal Cells" Molecules 22, no. 10: 1793. https://doi.org/10.3390/molecules22101793
APA StyleYang, F., He, C., Sun, X., Wang, J., Luo, C., Liu, G., Yang, L., Xiong, J., & Huo, L. (2017). The Regulatory Mechanism of MLT/MT1 Signaling on the Growth of Antler Mesenchymal Cells. Molecules, 22(10), 1793. https://doi.org/10.3390/molecules22101793