Bacterial Expression of Human Butyrylcholinesterase as a Tool for Nerve Agent Bioscavengers Development
Abstract
:1. Introduction
2. Results
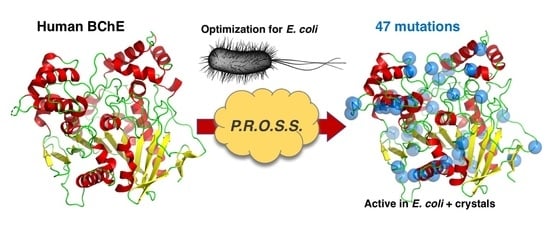
2.1. PROSS Processing
2.2. Constructs Cloning and Expression Screen
2.3. Purification of hBChE-7
2.4. Enzymatic Characterization
2.5. Oligomeric State Characterization
2.6. Protein Stability and Further Stabilization
2.7. hBChE-7 Crystallization
2.8. hBChE-7 X-ray Structure
3. Discussion
4. Materials and Methods
4.1. PROSS Processing
4.2. BChE Constructs
4.3. Expression and Purification
4.4. BChE Activity
4.5. SEC-MALS Analysis
4.6. Differential Scanning Fluorimetry Assay
4.7. Crystallogenesis, X-ray Data Collection and Analysis
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Lenz, D.E.; Maxwell, D.M.; Koplovitz, I.; Clark, C.R.; Capacio, B.R.; Cerasoli, D.M.; Federko, J.M.; Luo, C.; Saxena, A.; Doctor, B.P.; et al. Protection against soman or VX poisoning by human butyrylcholinesterase in guinea pigs and cynomolgus monkeys. Chem. Biol. Interact. 2005, 157–158, 205–210. [Google Scholar] [CrossRef] [PubMed]
- Carmona, G.N.; Jufer, R.A.; Goldberg, S.R.; Gorelick, D.A.; Greig, N.H.; Yu, Q.S.; Cone, E.J.; Schindler, C.W. Butyrylcholinesterase accelerates cocaine metabolism: In vitro and in vivo effects in nonhuman primates and humans. Drug Metab. Dispos. 2000, 28, 367–371. [Google Scholar] [PubMed]
- Brimijoin, S.; Chen, V.P.; Pang, Y.P.; Geng, L.; Gao, Y. Physiological roles for butyrylcholinesterase: A bche-ghrelin axis. Chem. Biol. Interact. 2016, 259, 271–275. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Yang, H.; Chen, Y.; Sun, H. Recent progress in the identification of selective butyrylcholinesterase inhibitors for alzheimer’s disease. Eur. J. Med. Chem. 2017, 132, 294–309. [Google Scholar] [CrossRef] [PubMed]
- Kosak, U.; Knez, D.; Coquelle, N.; Brus, B.; Pislar, A.; Nachon, F.; Brazzolotto, X.; Kos, J.; Colletier, J.P.; Gobec, S. N-propargylpiperidines with naphthalene-2-carboxamide or naphthalene-2-sulfonamide moieties: Potential multifunctional anti-alzheimer’s agents. Bioorg. Med. Chem. 2017, 25, 633–645. [Google Scholar] [CrossRef] [PubMed]
- Brazzolotto, X.; Wandhammer, M.; Ronco, C.; Trovaslet, M.; Jean, L.; Lockridge, O.; Renard, P.Y.; Nachon, F. Human butyrylcholinesterase produced in insect cells: Huprine-based affinity purification and crystal structure. FEBS J. 2012, 279, 2905–2916. [Google Scholar] [CrossRef] [PubMed]
- Masson, P.; Adkins, S.; Gouet, P.; Lockridge, O. Recombinant human butyrylcholinesterase G390V, the fluoride-2 variant, expressed in chinese hamster ovary cells, is a low affinity variant. J. Biol. Chem. 1993, 268, 14329–14341. [Google Scholar] [PubMed]
- Nachon, F.; Nicolet, Y.; Viguie, N.; Masson, P.; Fontecilla-Camps, J.C.; Lockridge, O. Engineering of a monomeric and low-glycosylated form of human butyrylcholinesterase: Expression, purification, characterization and crystallization. Eur. J. Biochem. 2002, 269, 630–637. [Google Scholar] [CrossRef] [PubMed]
- Terekhov, S.; Smirnov, I.; Bobik, T.; Shamborant, O.; Zenkova, M.; Chernolovskaya, E.; Gladkikh, D.; Murashev, A.; Dyachenko, I.; Palikov, V.; et al. A novel expression cassette delivers efficient production of exclusively tetrameric human butyrylcholinesterase with improved pharmacokinetics for protection against organophosphate poisoning. Biochimie 2015, 118, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Nachon, F.; Brazzolotto, X.; Trovaslet, M.; Masson, P. Progress in the development of enzyme-based nerve agent bioscavengers. Chem. Biol. Interact. 2013, 206, 536–544. [Google Scholar] [CrossRef] [PubMed]
- Ashani, Y.; Pistinner, S. Estimation of the upper limit of human butyrylcholinesterase dose required for protection against organophosphates toxicity: A mathematically based toxicokinetic model. Toxicol. Sci. 2004, 77, 358–367. [Google Scholar] [CrossRef] [PubMed]
- Saxena, A.; Tipparaju, P.; Luo, C.; Doctor, B.P. Pilot-scale production of human serum butyrylcholinesterase suitable for use as a bioscanvenger against nerve agent toxicity. Process Biochem. 2010, 45, 1313–1318. [Google Scholar] [CrossRef]
- Lockridge, O.; Schopfer, L.M.; Winger, G.; Woods, J.H. Large scale purification of butyrylcholinesterase from human plasma suitable for injection into monkeys; a potential new therapeutic for protection against cocaine and nerve agent toxicity. J. Med. Chem. Biol. Radiol. Def. 2005, 3, nihms5095. [Google Scholar] [PubMed]
- Saxena, A.; Sun, W.; Luo, C.; Doctor, B.P. Human serum butyrylcholinesterase: In vitro and in vivo stability, pharmacokinetics, and safety in mice. Chem. Biol. Interact. 2005, 157–158, 199–203. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Doctor, B.P.; Saxena, A. Safety and pharmacokinetics of human serum butyrylcholinesterase in guinea pigs. Chem. Biol. Interact. 2005, 157–158, 428–429. [Google Scholar] [CrossRef] [PubMed]
- Geyer, B.C.; Kannan, L.; Garnaud, P.E.; Broomfield, C.A.; Cadieux, C.L.; Cherni, I.; Hodgins, S.M.; Kasten, S.A.; Kelley, K.; Kilbourne, J.; et al. Plant-derived human butyrylcholinesterase, but not an organophosphorous-compound hydrolyzing variant thereof, protects rodents against nerve agents. Proc. Natl. Acad. Sci. USA 2010, 107, 20251–20256. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.J.; Huang, Y.; Baldassarre, H.; Wang, B.; Lazaris, A.; Leduc, M.; Bilodeau, A.S.; Bellemare, A.; Cote, M.; Herskovits, P.; et al. Recombinant human butyrylcholinesterase from milk of transgenic animals to protect against organophosphate poisoning. Proc. Natl. Acad. Sci. USA 2007, 104, 13603–13608. [Google Scholar] [CrossRef] [PubMed]
- Pope, C.; Uchea, C.; Flynn, N.; Poindexter, K.; Geng, L.; Brimijoin, W.S.; Hartson, S.; Ranjan, A.; Ramsey, J.D.; Liu, J. In vitro characterization of cationic copolymer-complexed recombinant human butyrylcholinesterase. Biochem. Pharmacol. 2015, 98, 531–539. [Google Scholar] [CrossRef] [PubMed]
- Masson, P.; Nachon, F. Cholinesterase reactivators and bioscavengers for pre- and post-exposure treatments of organophosphorus poisoning. J. Neurochem. 2017, 142 (Suppl. 2), 26–40. [Google Scholar] [CrossRef] [PubMed]
- Kolarich, D.; Weber, A.; Pabst, M.; Stadlmann, J.; Teschner, W.; Ehrlich, H.; Schwarz, H.P.; Altmann, F. Glycoproteomic characterization of butyrylcholinesterase from human plasma. Proteomics 2008, 8, 254–263. [Google Scholar] [CrossRef] [PubMed]
- Goldenzweig, A.; Goldsmith, M.; Hill, S.E.; Gertman, O.; Laurino, P.; Ashani, Y.; Dym, O.; Unger, T.; Albeck, S.; Prilusky, J.; et al. Automated structure- and sequence-based design of proteins for high bacterial expression and stability. Mol. Cell 2016, 63, 337–346. [Google Scholar] [CrossRef] [PubMed]
- Lobstein, J.; Emrich, C.A.; Jeans, C.; Faulkner, M.; Riggs, P.; Berkmen, M. Shuffle, a novel Escherichia coli protein expression strain capable of correctly folding disulfide bonded proteins in its cytoplasm. Microb. Cell Fact. 2012, 11, 56. [Google Scholar] [CrossRef] [PubMed]
- Phan, J.; Zdanov, A.; Evdokimov, A.G.; Tropea, J.E.; Peters, H.K., 3rd; Kapust, R.B.; Li, M.; Wlodawer, A.; Waugh, D.S. Structural basis for the substrate specificity of tobacco etch virus protease. J. Biol. Chem. 2002, 277, 50564–50572. [Google Scholar] [CrossRef] [PubMed]
- Radic, Z.; Pickering, N.A.; Vellom, D.C.; Camp, S.; Taylor, P. Three distinct domains in the cholinesterase molecule confer selectivity for acetyl- and butyrylcholinesterase inhibitors. Biochemistry 1993, 32, 12074–12084. [Google Scholar] [CrossRef] [PubMed]
- Lockridge, O.; Blong, R.M.; Masson, P.; Froment, M.T.; Millard, C.B.; Broomfield, C.A. A single amino acid substitution, Gly117His, confers phosphotriesterase (organophosphorus acid anhydride hydrolase) activity on human butyrylcholinesterase. Biochemistry 1997, 36, 786–795. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Schopfer, L.M.; Masson, P.; Lockridge, O. Lamellipodin proline rich peptides associated with native plasma butyrylcholinesterase tetramers. Biochem. J. 2008, 411, 425–432. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.; Schopfer, L.M.; Lockridge, O. Origin of polyproline-rich peptides in human butyrylcholinesterase tetramers. Chem. Biol. Interact. 2016, 259, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Schopfer, L.M.; Delacour, H.; Masson, P.; Leroy, J.; Krejci, E.; Lockridge, O. The C5 variant of the butyrylcholinesterase tetramer includes a noncovalently bound 60 kDa lamellipodin fragment. Molecules 2017, 22, 1083. [Google Scholar] [CrossRef] [PubMed]
- Cheung, J.; Rudolph, M.J.; Burshteyn, F.; Cassidy, M.S.; Gary, E.N.; Love, J.; Franklin, M.C.; Height, J.J. Structures of human acetylcholinesterase in complex with pharmacologically important ligands. J. Med. Chem. 2012, 55, 10282–10286. [Google Scholar] [CrossRef] [PubMed]
- Bourne, Y.; Taylor, P.; Radic, Z.; Marchot, P. Structural insights into ligand interactions at the acetylcholinesterase peripheral anionic site. EMBO J. 2003, 22, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Raves, M.L.; Harel, M.; Pang, Y.P.; Silman, I.; Kozikowski, A.P.; Sussman, J.L. Structure of acetylcholinesterase complexed with the nootropic alkaloid, (-)-huperzine A. Nat. Struct. Biol. 1997, 4, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Dighe, S.N.; Deora, G.S.; De la Mora, E.; Nachon, F.; Chan, S.; Parat, M.O.; Brazzolotto, X.; Ross, B.P. Discovery and structure-activity relationships of a highly selective butyrylcholinesterase inhibitor by structure-based virtual screening. J. Med. Chem. 2016, 59, 7683–7689. [Google Scholar] [CrossRef] [PubMed]
- Zeldin, O.B.; Gerstel, M.; Garman, E.F. Raddose-3d: Time- and space-resolved modelling of dose in macromolecular crystallography. J. Appl. Crystallogr. 2013, 46, 1225–1230. [Google Scholar] [CrossRef]
- Masson, P.; Legrand, P.; Bartels, C.F.; Froment, M.T.; Schopfer, L.M.; Lockridge, O. Role of aspartate 70 and tryptophan 82 in binding of succinyldithiocholine to human butyrylcholinesterase. Biochemistry 1997, 36, 2266–2277. [Google Scholar] [CrossRef] [PubMed]
- Masson, P.; Xie, W.; Froment, M.T.; Lockridge, O. Effects of mutations of active site residues and amino acids interacting with the omega loop on substrate activation of butyrylcholinesterase. Biochim. Biophys. Acta 2001, 1544, 166–176. [Google Scholar] [CrossRef]
- Masson, P.; Froment, M.T.; Bartels, C.F.; Lockridge, O. Asp7O in the peripheral anionic site of human butyrylcholinesterase. Eur. J. Biochem. 1996, 235, 36–48. [Google Scholar] [CrossRef] [PubMed]
- Duysen, E.G.; Bartels, C.F.; Lockridge, O. Wild-type and A328W mutant human butyrylcholinesterase tetramers expressed in Chinese hamster ovary cells have a 16-hour half-life in the circulation and protect mice from cocaine toxicity. J. Pharmacol. Exp. Ther. 2002, 302, 751–758. [Google Scholar] [CrossRef] [PubMed]
- Jevsevar, S.; Kunstelj, M.; Porekar, V.G. Pegylation of therapeutic proteins. Biotechnol. J. 2010, 5, 113–128. [Google Scholar] [CrossRef] [PubMed]
- Goldsmith, M.; Aggarwal, N.; Ashani, Y.; Jubran, H.; Greisen, P.J.; Ovchinnikov, S.; Leader, H.; Baker, D.; Sussman, J.L.; Goldenzweig, A.; et al. Overcoming an optimization plateau in the directed evolution of highly efficient nerve agent bioscavengers. Protein Eng. Des. Sel. 2017, 30, 333–345. [Google Scholar] [CrossRef] [PubMed]
- Millard, C.B.; Lockridge, O.; Broomfield, C.A. Design and expression of organophosphorus acid anhydride hydrolase activity in human butyrylcholinesterase. Biochemistry 1995, 34, 15925–15933. [Google Scholar] [CrossRef] [PubMed]
- Millard, C.B.; Lockridge, O.; Broomfield, C.A. Organophosphorus acid anhydride hydrolase activity in human butyrylcholinesterase: Synergy results in a somanase. Biochemistry 1998, 37, 237–247. [Google Scholar] [CrossRef] [PubMed]
- Zheng, F.; Xue, L.; Hou, S.; Liu, J.; Zhan, M.; Yang, W.; Zhan, C.G. A highly efficient cocaine-detoxifying enzyme obtained by computational design. Nat. Commun. 2014, 5, 3457. [Google Scholar] [CrossRef] [PubMed]
- Katalinic, M.; Macek Hrvat, N.; Baumann, K.; Morasi Pipercic, S.; Makaric, S.; Tomic, S.; Jovic, O.; Hrenar, T.; Milicevic, A.; Jelic, D.; et al. A comprehensive evaluation of novel oximes in creation of butyrylcholinesterase-based nerve agent bioscavengers. Toxicol. Appl. Pharmacol. 2016, 310, 195–204. [Google Scholar] [CrossRef] [PubMed]
- Kosak, U.; Brus, B.; Knez, D.; Sink, R.; Zakelj, S.; Trontelj, J.; Pislar, A.; Slenc, J.; Gobec, M.; Zivin, M.; et al. Development of an in vivo active reversible butyrylcholinesterase inhibitor. Sci. Rep. 2016, 6, 39495. [Google Scholar] [CrossRef] [PubMed]
- Chiu, J.; March, P.E.; Lee, R.; Tillett, D. Site-directed, ligase-independent mutagenesis (slim): A single-tube methodology approaching 100% efficiency in 4 h. Nucleic Acids Res. 2004, 32, e174. [Google Scholar] [CrossRef] [PubMed]
- Ellman, G.L.; Courtney, K.D.; Andres, V., Jr.; Feather-Stone, R.M. A new and rapid colorimetric determination of acetylcholinesterase activity. Biochem. Pharmacol. 1961, 7, 88–95. [Google Scholar] [CrossRef]
- Boivin, S.; Kozak, S.; Meijers, R. Optimization of protein purification and characterization using thermofluor screens. Protein Expr. Purif. 2013, 91, 192–206. [Google Scholar] [CrossRef] [PubMed]
- De Sanctis, D.; Beteva, A.; Caserotto, H.; Dobias, F.; Gabadinho, J.; Giraud, T.; Gobbo, A.; Guijarro, M.; Lentini, M.; Lavault, B.; et al. Id29: A high-intensity highly automated esrf beamline for macromolecular crystallography experiments exploiting anomalous scattering. J. Synchrotron Radiat. 2012, 19, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Kabsch, W. XDS. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Monaco, S.; Gordon, E.; Bowler, M.W.; Delageniere, S.; Guijarro, M.; Spruce, D.; Svensson, O.; McSweeney, S.M.; McCarthy, A.A.; Leonard, G.; et al. Automatic processing of macromolecular crystallography X-ray diffraction data at the esrf. J. Appl. Crystallogr. 2013, 46, 804–810. [Google Scholar] [CrossRef] [PubMed]
- Adams, P.D.; Afonine, P.V.; Bunkoczi, G.; Chen, V.B.; Davis, I.W.; Echols, N.; Headd, J.J.; Hung, L.W.; Kapral, G.J.; Grosse-Kunstleve, R.W.; et al. Phenix: A comprehensive python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 213–221. [Google Scholar] [CrossRef] [PubMed]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of coot. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 486–501. [Google Scholar] [CrossRef] [PubMed]
- Ho, B.K.; Gruswitz, F. Hollow: Generating accurate representations of channel and interior surfaces in molecular structures. BMC Struct. Biol. 2008, 8, 49. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Not available. |







| Enzymatic Activity | Dimer Interface |
|---|---|
| 67, 68, 70, 77, 81, 82,83, 84, 112, 114, 115, 116, 117, 118, 119, 120, 128, 146, 197, 198, 199, 224, 231, 276, 277, 285, 286, 287, 288, 289, 322, 325, 328, 329, 332, 398, 430, 433, 434, 437, 438, 439, 440, 441, 442 | 364, 367, 371, 372, 517, 520, 521, 525, 528 |
| Name | Mutations Number | Mutation Positions |
|---|---|---|
| hBChE-1 | 9 | 7, 48, 54, 215, 250, 397, 454, 466, 468 |
| hBChE-2 | 15 | 7, 48, 54, 62, 110, 215, 236, 237, 250, 397, 406, 412, 454, 466, 468 |
| hBChE-3 | 18 | 7, 48, 54, 110, 126, 180, 215, 236, 237, 250, 274, 379, 397, 406, 412, 454, 466, 468 |
| hBChE-4 | 21 | 7, 48, 54, 110, 126, 180, 215, 227, 236, 237, 250, 274, 360, 379, 397, 406, 412, 454, 466, 468, 469 |
| hBChE-5 | 25 | 7, 48, 54, 110, 111, 126, 180, 215, 227, 236, 237, 250, 274, 360, 379, 397, 406, 409, 412, 454, 466, 468, 469, 489, 523 |
| hBChE-6 | 36 | 7, 48, 54, 66, 71, 110, 111, 126, 176, 180, 215, 227, 234, 236, 237, 250, 274, 305, 356, 360, 377, 379, 380, 391, 397, 406, 409, 412, 417, 454, 466, 468, 469, 489, 518, 523 |
| hBChE-7 | 47 | 7, 48, 54, 66, 71, 110, 111, 126, 176, 180, 188, 190, 191, 215, 227, 234, 236, 237, 250, 274, 283, 305, 342, 356, 360, 377, 379, 380, 387, 390, 391, 397, 406, 409, 410, 412, 417, 454, 459, 466, 468, 469, 489, 495, 508, 518, 523 |
| Ks (µM) | Kss (µM) | b | kcat (min−1) | |
|---|---|---|---|---|
| hBChE-7 | 30.0 ± 2.5 | 1291 ± 112 | 2.80 ± 0.10 | 46,715 |
| hBChECHO [8] | 25.6 ± 0.4 | 510 ± 35 | 2.85 ± 0.15 | 28,000 |
| hBChEplasma [25] | 20 | 300 | 2.4 | 24,000 |
| hBChE-7 | |
|---|---|
| Data collection | |
| X-ray source—beamline | ESRF—ID29-1 |
| Wavelength (Å) | 1.074 |
| Resolution range (Å) (highest resolution shell) | 65.02–2.476 (2.565–2.476) |
| Space group | C 1 2 1 |
| Unit cell parameters (Å) ° | 159.9 75.1 122.1 90.0 93.4 90.0 |
| Total reflections | 175,981 (17,785) |
| Unique reflections | 51,072 (5027) |
| Multiplicity | 3.4 (3.5) |
| Completeness (%) | 98.37 (97.86) |
| Mean I/sigma | 7.92 (2.30) |
| Wilson B-factor | 36.13 |
| R-merge | 0.1163 (0.5303) |
| R-meas | 0.1381 (0.6246) |
| R-pim | 0.0737 (0.3278) |
| CC1/2 | 0.995 (0.887) |
| CC * | 0.999 (0.969) |
| Refinement statistics | |
| Reflections used in refinement | 50,925 (5021) |
| Reflections used for R-free | 2574 (241) |
| R-work | 0.2219 (0.2937) |
| R-free | 0.2553 (0.3305) |
| CC (work) | 0.956 (0.880) |
| CC (free) | 0.947 (0.831) |
| Number of non-hydrogen atoms | 9018 |
| Macromolecules | 8423 |
| Ligands | 219 |
| Solvent | 376 |
| Protein residues | 1054 |
| RMS (bonds) | 0.003 |
| RMS (angles) | 0.54 |
| Ramachandran favored (%) | 95.62 |
| Ramachandran allowed (%) | 4.00 |
| Ramachandran outliers (%) | 0.38 |
| Clashscore | 5.66 |
| Average B-factor | 41.57 |
| macromolecule | 41.08 |
| ligands | 56.33 |
| solvent | 44.16 |
| Name | Sequence |
|---|---|
| 8His-F1 | 5′-CATCACCATCACCATCACCATCACTCTGATAAAATTATTCATCTG-3′ |
| 8His-F2 | 5′-TCTGATAAAATTATTCATCTG-3′ |
| 8His-R1 | 5′-GTGATGGTGATGGTGATGGTGATGCATATGTATATACCTCTTTAA-3′ |
| 8His-R2 | 5′-CATATGTATATACCTCTTTAA-3′ |
| TEV-F1 | 5′-GAGAATCTTTATTTTCAGGGCGCCATGGAAGATGACATTATCATC-3′ |
| TEV-F2 | 5′-CCATGGAAGATGACATTATCATC-3′ |
| TEV-R1 | 5′-CGCCCTGAAAATAAAGATTCTCACCGGATCCAGAGCCGGCCAG-3′ |
| TEV-R2 | 5′-ACCGGATCCAGAGCCGGCCAG-3′ |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brazzolotto, X.; Igert, A.; Guillon, V.; Santoni, G.; Nachon, F. Bacterial Expression of Human Butyrylcholinesterase as a Tool for Nerve Agent Bioscavengers Development. Molecules 2017, 22, 1828. https://doi.org/10.3390/molecules22111828
Brazzolotto X, Igert A, Guillon V, Santoni G, Nachon F. Bacterial Expression of Human Butyrylcholinesterase as a Tool for Nerve Agent Bioscavengers Development. Molecules. 2017; 22(11):1828. https://doi.org/10.3390/molecules22111828
Chicago/Turabian StyleBrazzolotto, Xavier, Alexandre Igert, Virginia Guillon, Gianluca Santoni, and Florian Nachon. 2017. "Bacterial Expression of Human Butyrylcholinesterase as a Tool for Nerve Agent Bioscavengers Development" Molecules 22, no. 11: 1828. https://doi.org/10.3390/molecules22111828
APA StyleBrazzolotto, X., Igert, A., Guillon, V., Santoni, G., & Nachon, F. (2017). Bacterial Expression of Human Butyrylcholinesterase as a Tool for Nerve Agent Bioscavengers Development. Molecules, 22(11), 1828. https://doi.org/10.3390/molecules22111828