Comparative Gene Expression Analysis in WM164 Melanoma Cells Revealed That β-β-Dimethylacrylshikonin Leads to ROS Generation, Loss of Mitochondrial Membrane Potential, and Autophagy Induction
Abstract
:1. Introduction
2. Results and Discussion
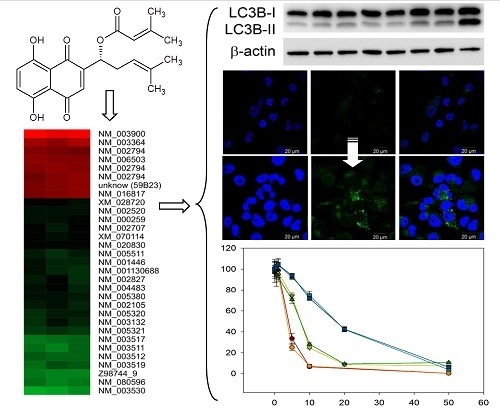
2.1. Comparative Gene Expression Analysis Revealed 31 Distinct mRNAs as at Least 2-Fold Significantly Differentially Expressed with Sequestosome 1 (p62) mRNA as Largest Change
2.2. DMAS Favors Catabolic Processes
2.3. DMAS Increased the Expression of the Autophagy-Associated Protein LC3B-II
2.4. DMAS Leads to ROS Generation and Loss of Mitochondrial Membrane Potential
2.5. Cytotoxicity of DMAS Against Non-Tumorigenic Cells Depends on Cell Type
3. Material and Methods
3.1. Isolation and Identification of DMAS
3.2. Cell Culture
3.3. Sample Preparation for Microarray-Based Transcription Profiling
3.4. RNA Extraction and RT-qPCR
3.5. Microarray-Based Transcription Profiling
3.6. Microarray Data Analysis
3.7. Western Blot
3.8. Immunofluorescence
3.9. ROS-Glo™ H2O2 Assay
3.10. CellTiter-Glo® Luminescent Cell Viability Assay
3.11. MITO-ID® Membrane Potential Detection Kit
3.12. XTT Assay
3.13. Statistical Analysis
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Skin Cancers. Available online: http://www.who.int/uv/faq/skincancer/en/index1.html (accessed on 28 September 2018).
- Singh, S.; Zafar, A.; Khan, S.; Naseem, I. Towards therapeutic advances in melanoma management: An overview. Life Sci. 2017, 174, 50–58. [Google Scholar] [CrossRef] [PubMed]
- Aamdal, S. Current approaches to adjuvant therapy of melanoma. Eur. J. Cancer 2011, 47 (Suppl. 3), 336–337. [Google Scholar] [CrossRef]
- Middleton, M.R.; Lee, S.M.; Arance, A.; Wood, M.; Thatcher, N.; Margison, G.P. O6-methylguanine formation, repair protein depletion and clinical outcome with a 4 h schedule of temozolomide in the treatment of advanced melanoma: Results of a phase II study. Int. J. Cancer 2000, 88, 469–473. [Google Scholar] [CrossRef]
- Bichakjian, C.K.; Halpern, A.C.; Johnson, T.M.; Foote Hood, A.; Grichnik, J.M.; Swetter, S.M.; Tsao, H.; Barbosa, V.H.; Chuang, T.Y.; Duvic, M.; et al. Guidelines of care for the management of primary cutaneous melanoma. American Society of Dermatology. J. Am. Acad. Dermatol. 2011, 65, 1032–1047. [Google Scholar] [CrossRef] [PubMed]
- Davies, H.; Bignell, G.R.; Cox, C.; Stephens, P.; Edkins, S.; Clegg, S.; Teague, J.; Woffendin, H.; Garnett, M.J.; Bottomley, W.; et al. Mutations of the BRAF gene in human cancer. Nature 2002, 417, 949–954. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smalley, K.S.; Haass, N.K.; Brafford, P.A.; Lioni, M.; Flaherty, K.T.; Herlyn, M. Multiple signaling pathways must be targeted to overcome drug resistance in cell lines derived from melanoma metastases. Mol. Cancer Ther. 2006, 5, 1136–1144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Newman, D.J.; Cragg, G.M. Natural products as sources of new drugs from 1981 to 2014. J. Nat. Prod. 2016, 79, 629–661. [Google Scholar] [CrossRef] [PubMed]
- Zhongzhen, Z. An illustrated Chinese Materia Medica in Hong Kong; Hong Kong School of Chinese Medicine: Hong Kong, China, 2004. [Google Scholar]
- The State Pharmacopoeia Commission of the People’s Republic of China. Pharmacopoeia of the People´s Republic of China, English ed.; Chemical Industry Press: Beijing, China, 2000. [Google Scholar]
- Duke, J.A.; Ayensu, S. Medicinal plants of China; Reference Publications Inc.: Algonac, MI, USA, 1985. [Google Scholar]
- Papageorgiou, V.P.; Assimopoulou, A.N.; Couladouros, E.A.; Hepworth, D.; Nicolaou, K.C. The chemistry and biology of alkannin, shikonin and related naphthazarin natural products. Angew. Chem. Int. Ed. 1999, 38, 270–301. [Google Scholar] [CrossRef]
- Chen, X.; Yang, L.; Oppenheim, J.J.; Howard, M.Z. Cellular pharmacology studies of shikonin derivatives. Phytother. Res. 2002, 16, 199–209. [Google Scholar] [CrossRef] [PubMed]
- Andújar, I.; Rios, J.L.; Giner, R.M.; Recio, M.C. Pharmacological properties of shikonin—A review of literature since 2002. Planta Med. 2013, 79, 1685–1697. [Google Scholar] [CrossRef] [PubMed]
- Rinner, B.; Kretschmer, N.; Knausz, H.; Mayer, A.; Boechzelt, H.; Hao, X.J.; Heubl, G.; Efferth, T.; Schaider, H.; Bauer, R.J. A petrol ether extract of the roots of Onosma paniculatum induces cell death in a caspase dependent manner. J. Ethnopharmacol. 2010, 129, 182–188. [Google Scholar] [CrossRef] [PubMed]
- Kretschmer, N.; Rinner, B.; Deutsch, A.J.; Lohberger, B.; Knausz, H.; Kunert, O.; Blunder, M.; Boechzelt, H.; Schaider, H.; Bauer, R. Naphthoquinones from Onosma paniculata induce cell-cycle arrest and apoptosis in melanoma Cells. J. Nat. Prod. 2012, 75, 865–869. [Google Scholar] [CrossRef] [PubMed]
- Moscat, J.; Diaz-Meco, M.T. P62: A versatile multitasker takes on cancer. Trends Biochem. Sci. 2012, 37, 230–236. [Google Scholar] [CrossRef] [PubMed]
- Kamburov, A.; Stelzl, U.; Lehrach, H.; Herwig, R. The ConsensusPathDB interaction database: 2013 update. Nucl. Acids Res. 2013, 41, 793–800. [Google Scholar] [CrossRef] [PubMed]
- Jiang, T.; Harder, B.; Rojo de la Vega, M.; Wong, P.K.; Chapman, E.; Zhang, D.D. P62 links autophagy and Nrf2 signaling. Free Radic. Biol. Med. 2015, 88, 199–204. [Google Scholar] [CrossRef] [PubMed]
- Kansanen, E.; Kuosmanen, S.M.; Leinonen, H.; Levonen, A.L. The Keap1-Nrf2 pathway: Mechanisms of activation and dysregulation in cancer. Redox. Biol. 2013, 1, 45–49. [Google Scholar] [CrossRef] [PubMed]
- Sporn, M.B.; Liby, K.T. NRF2 and cancer: The good, the bad and the importance of context. Nat. Rev. Cancer 2012, 12, 564–571. [Google Scholar] [CrossRef] [PubMed]
- Probst, B.L.; McCauley, L.; Trevino, I.; Wigley, W.C.; Ferguson, D.A. Cancer cell growth is differentially affected by constitutive activation of NRF2 by KEAP1 deletion and pharmacological activation of NRF2 by the synthetic triterpenoid, RTA 405. PLoS ONE 2015, 10, e0135257. [Google Scholar] [CrossRef] [PubMed]
- Liby, K.T. Synthetic triterpenoids can protect against toxicity without reducing the efficacy of treatment with carboplatin and paclitaxel in experimental lung cancer. Dose Response 2013, 12, 136–151. [Google Scholar] [CrossRef] [PubMed]
- Hwang, B.J.; Adhikary, G.; Eckert, R.L.; Lu, A.L. Chk1 inhibition as a novel therapeutic strategy in melanoma. Oncotarget 2018, 9, 30450–30464. [Google Scholar] [CrossRef] [PubMed]
- Satyamoorthy, K.; Li, G.; Gerrero, M.R.; Brose, M.S.; Volpe, P.; Weber, B.L.; van Belle, P.; Elder, D.E.; Herlyn, M. Constitutive Mitogen-activated Protein Kinase Activation in Melanoma Is Mediated by Both BRAF Mutations and Autocrine Growth Factor Stimulation. Cancer Res. 2003, 63, 756–759. [Google Scholar] [PubMed]
- Mizushima, N.; Yoshimori, T. How to interpret LC3 immunoblotting. Autophagy 2007, 3, 542–545. [Google Scholar] [CrossRef] [PubMed]
- Corazzari, M.; Rapino, F.; Ciccosanti, F.; Giglio, P.; Antonioli, M.; Conti, B.; Fimia, G.M.; Lovat, P.E.; Piacentini, M. Oncogenic BRAF induces chronic ER stress condition resulting in increased basal autophagy and apoptotic resistance of cutaneous melanoma. Cell Death Diff. 2015, 22, 946–958. [Google Scholar] [CrossRef] [PubMed]
- Kraya, A.A.; Piao, S.; Xu, X.; Zhang, G.; Herlyn, M.; Gimotty, P.; Levine, B.; Amaravadi, R.K.; Speicher, D.W. Identification of secreted proteins that reflect autophagy dynamics within tumor cells. Autophagy 2015, 11, 60–74. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.J.; Wang, Y.; Chen, H.Z.; Xing, Y.Z.; Li, F.W.; Zhang, Q.; Zhou, B.; Zhang, H.K.; Zhang, J.; Bian, X.L.; et al. Orphan nuclear receptor TR3 acts in autophagic cell death via mitochondrial signaling pathway. Nat. Chem. Biol. 2014, 10, 133–140. [Google Scholar] [CrossRef] [PubMed]
- Al-Qatati, A.; Aliwaini, S. Combined pitavastatin and dacarbazine treatment activates apoptosis and autophagy resulting in synergistic cytotoxicity in melanoma cells. Oncol. Lett. 2017, 14, 7993–7999. [Google Scholar] [CrossRef] [PubMed]
- Poillet-Perez, L.; Despouy, G.; Delage-Mourroux, R.; Boyer-Guittaut, M. Interplay between ROS and autophagy in cancer cells, from tumor initiation to cancer therapy. Redox. Biol. 2015, 4, 184–192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Z.Y.; Yang, Y.; Ming, M.; Liu, B. Mitochondrial ROS generation for regulation of autophagic pathways in cancer. Biochem. Biophys. Res. Commun. 2011, 414, 5–8. [Google Scholar] [CrossRef] [PubMed]
- Halliwell, B. Oxidative stress in cell culture: An under-appreciated problem? FEBS Lett. 2003, 540, 3–6. [Google Scholar] [CrossRef]
- Babich, H.; Ackerman, N.J.; Burekhovich, F.; Zuckerbraun, H.L.; Schuck, A.G. Gingko biloba leaf extract induces oxidative stress in carcinoma HSC-2 cells. Toxicol. In Vitro 2009, 23, 992–999. [Google Scholar] [CrossRef] [PubMed]
- Kelts, J.L.; Cali, J.J.; Duellman, S.J.; Shultz, J. Altered cytotoxicity of ROS-inducing compounds by sodium pyruvate in cell culture medium depends on the location of ROS generation. SpringerPlus 2015, 4, 269. [Google Scholar] [CrossRef] [PubMed]
- Thor, H.; Smith, M.T.; Hartzell, P.; Bellomo, G.; Jewell, S.A.; Orrenius, S. The metabolism of menadione (2-methyl-1,4-naphthoquione) by isolated hepatocytes. A study of the implications of oxidative stress in intact cells. J. Biol. Chem. 1982, 257, 12419–12425. [Google Scholar] [PubMed]
- Kwak, S.Y.; Jeong, Y.K.; Kim, B.Y.; Lee, J.Y.; Ahn, H.J.; Jeong, J.H.; Kim, M.S.; Kim, J.; Han, Y.H. Dimethylacrylshikonin sensitizes human colon cancer cells to ionizing radiation through the upregulation of reactive oxygen species. Oncol. Lett. 2014, 7, 1812–1818. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Lam, G.Y.; Brumell, J.H. Autophagy signaling through reactive oxygen species. Antiox. Redox. Signal 2011, 14, 2215–2231. [Google Scholar] [CrossRef] [PubMed]
- Wiench, B.; Eichhorn, T.; Paulsen, M.; Efferth, T. Shikonin directly targets mitochondria and causes mitochondrial dysfunction in cancer cells. Evid.-Based Complement. Altern. Med. 2012, 2012, 726025. [Google Scholar] [CrossRef] [PubMed]
- Gara, R.K.; Srivastava, V.K.; Duggal, S.; Bagga, J.K.; Bhatt, M.L.B.; Sanyal, S.; Mishra, D.P. Shikonin selectively induces apoptosis in human prostate cancer cells through the endoplasmic reticulum stress and mitochondrial apoptotic pathway. J. Biomed. Sci. 2015, 22, 26. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Kang, K.A.; Piao, M.J.; Zhen, A.X.; Hyun, Y.J.; Kim, H.M.; Ryu, Y.S.; Hyun, J.W. Shikonin Exerts Cytotoxic Effects in Human Colon Cancers by Inducing Apoptotic Cell Death via the Endoplasmic Reticulum and Mitochondria-Mediated Pathways. Biomol. Ther. 2018, 7. [Google Scholar] [CrossRef] [PubMed]
- Liang, W.; Cai, A.; Chen, G.; Xi, H.; Wu, X.; Cui, J.; Zhang, K.; Zhao, X.; Yu, J.; Wei, B.; et al. Shikonin induces mitochondria-mediated apoptosis and enhances chemotherapeutic sensitivity of gastric cancer through reactive oxygen species. Sci. Rep. 2016, 6, 38267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, H.Y.; Chi, J.T.; Dudoit, S.; Bondre, D.; Van de Rijn, M.; Botstein, D.; Brown, P.O. Diversity, topographic differentiation, and positional memory in human fibroblasts. Proc. Natl. Acad. Sci. USA 2002, 99, 12877–12882. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, W.J.; Zhang, X.; Cheng, C.; Wang, F.; Wang, X.K.; Liang, Y.J.; To, K.K.W.; Zhou, W.; Huang, H.B.; Fu, L.W. Crizotinib (PF-02341066) reverses multidrug resistance in cancer cells by inhibiting the function of P.-glycoprotein. Br. J. Pharmacol. 2012, 166, 1669–1683. [Google Scholar] [CrossRef] [PubMed]
- Akiyode, O.; George, D.; Getti, G.; Boateng, J. Systematic comparison of the functional physico-chemical characteristics and biocidal activity of microbial derived biosurfactants on blood-derived and breast cancer cells. J. Colloid Interface Sci. 2016, 479, 221–233. [Google Scholar] [CrossRef] [PubMed]
- Peer, D.; Karp, J.M.; Hong, S.; Farokhzad, O.C.; Margalit, R.; Langer, R. Nanocarriers as an emerging platform for cancer therapy. Nat. Nanotechnol. 2007, 2, 751–760. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Tong, Y.; Bai, L.; Ye, L.; Zhong, L.; Duan, X.; Zhu, Y. Lactoferrin functionalized PEG-PLGA nanoparticles of shikonin for brain targeting therapy of glioma. Int. J. Biol. Macromol. 2018, 107, 204–211. [Google Scholar] [CrossRef] [PubMed]
- Kaur, A.; Jyoti, K.; Baldi, A.; Jain, U.K.; Chandra, R. Self-assembled nanomicelles of amphiphilic clotrimazole glycyl-glycine analogue augmented drug delivery, apoptosis and restrained melanoma tumour progression. Mater. Sci. Eng. C Mater. Biol. Appl. 2018, 89, 75–86. [Google Scholar] [CrossRef] [PubMed]
- Karbiener, M.; Pisani, D.F.; Frontini, A.; Oberreiter, L.M.; Lang, E.; Vegiopoulos, A.; Mössenböck, K.; Bernhardt, G.A.; Mayr, T.; Hildner, F.; et al. MicroRNA-26 family is required for human adipogenesis and drives characteristics of brown adipocytes. Stem Cells 2014, 32, 1578–1590. [Google Scholar] [CrossRef] [PubMed]
- Pieler, R.; Sanchez-Cabo, F.; Hackl, H.; Thallinger, G.G.; Trajanoski, Z. ArrayNorm: Comprehensive normalization and analysis of microarray data. Bioinformatics 2004, 20, 1971–1973. [Google Scholar] [CrossRef] [PubMed]
- Sturn, A.; Quackenbush, J.; Trajanoski, Z. Genesis: Cluster analysis of microarray data. Bioinformatics 2002, 18, 207–208. [Google Scholar] [CrossRef] [PubMed]
- Gene Ontology Consortium. Gene Ontology Consortium: Going forward. Nucleic Acids Res. 2015, 43, D1049–D1056. [Google Scholar] [CrossRef] [PubMed]








| Pathway | Set Size | Candidates | p-Value | Q-Value |
|---|---|---|---|---|
| NRF2 pathway | 142 | 3 (2.1%) | 0.0004 | 0.0057 |
| Proteasome-Homo sapiens (human) | 44 | 2 (4.5%) | 0.0009 | 0.0057 |
| Apoptosis-related network due to altered Notch3 in ovarian cancer | 53 | 2 (3.8%) | 0.0013 | 0.0057 |
| TLR JNK | 63 | 2 (3.2%) | 0.0018 | 0.0057 |
| IL-1 JNK | 63 | 2 (3.2%) | 0.0018 | 0.0057 |
| TLR p38 | 64 | 2 (3.1%) | 0.0019 | 0.0057 |
| Proteasome Degradation | 64 | 2 (3.1%) | 0.0019 | 0.0057 |
| DroToll-like | 65 | 2 (3.1%) | 0.0019 | 0.0057 |
| IL-1 NFkB | 65 | 2 (3.1%) | 0.0019 | 0.0057 |
| IL-1 p38 | 67 | 2 (3.0%) | 0.0021 | 0.0057 |
| TNF | 67 | 2 (3.0%) | 0.0021 | 0.0057 |
| TLR NFkB | 70 | 2 (2.9%) | 0.0022 | 0.0057 |
| Hedgehog | 72 | 2 (2.8%) | 0.0024 | 0.0057 |
| Notch | 79 | 2 (2.6%) | 0.0028 | 0.0062 |
| Nuclear Receptors Meta-Pathway | 316 | 3 (0.9%) | 0.0037 | 0.0076 |
| CD4 T cell receptor signaling-NFkB cascade | 95 | 2 (2.1%) | 0.0041 | 0.0080 |
| UCH proteinases | 107 | 2 (1.9%) | 0.0051 | 0.0093 |
| TGF-beta super family signaling pathway canonical | 115 | 2 (1.7%) | 0.0060 | 0.0099 |
| Wnt Canonical | 120 | 2 (1.7%) | 0.0064 | 0.0099 |
| Wnt Mammals | 120 | 2 (1.7%) | 0.0064 | 0.0099 |
| CD4 T cell receptor signaling | 128 | 2 (1.6%) | 0.0074 | 0.0109 |
| B cell receptor signaling | 134 | 2 (1.5%) | 0.0080 | 0.0113 |
| Pathway | Set Size | Candidates | p-Value | Q-Value |
|---|---|---|---|---|
| Metabolism of amino acids and derivatives | 328 | 5 (1.5%) | 0.0008 | 0.0086 |
| Nucleotide Metabolism | 19 | 2 (10.5%) | 0.0008 | 0.0086 |
| S-Adenosylhomocysteine (SAH) Hydrolase Deficiency | 20 | 2 (10.0%) | 0.0009 | 0.0086 |
| Methionine Metabolism | 20 | 2 (10.0%) | 0.0009 | 0.0086 |
| Methionine Adenosyltransferase Deficiency | 20 | 2 (10.0%) | 0.0009 | 0.0086 |
| Glycine N-methyltransferase Deficiency | 20 | 2 (10.0%) | 0.0009 | 0.0086 |
| Hypermethioninemia | 20 | 2 (10.0%) | 0.0009 | 0.0086 |
| Methylenetetrahydrofolate Reductase Deficiency (MTHFRD) | 20 | 2 (10.0%) | 0.0009 | 0.0086 |
| Homocystinuria-megaloblastic anemia due to defect in cobalamin metabolism, cblG complementation type | 20 | 2 (10.0%) | 0.0009 | 0.0086 |
| Cystathionine Beta-Synthase Deficiency | 20 | 2 (10.0%) | 0.0009 | 0.0086 |
| Methionine De Novo and Salvage Pathway | 21 | 2 (9.5%) | 0.0010 | 0.0087 |
| One Carbon Metabolism | 28 | 2 (7.1%) | 0.0019 | 0.0142 |
| Trans-sulfuration and one carbon metabolism | 31 | 2 (6.5%) | 0.0022 | 0.0158 |
| Thiopurine Pathway, Pharmacokinetics/Pharmacodynamics | 32 | 2 (6.2%) | 0.0024 | 0.0158 |
| TGF-beta receptor signaling activates SMADs | 34 | 2 (5.9%) | 0.0027 | 0.0167 |
| Metalloprotease DUBs | 37 | 2 (5.4%) | 0.0032 | 0.0176 |
| Selenoamino acid metabolism | 132 | 3 (2.3%) | 0.0032 | 0.0176 |
| Metabolism | 2035 | 11 (0.5%) | 0.0035 | 0.0181 |
| Cysteine and methionine metabolism-Homo sapiens (human) | 45 | 2 (4.4%) | 0.0047 | 0.0229 |
| The citric acid (TCA) cycle and respiratory electron transport | 171 | 3 (1.8%) | 0.0067 | 0.0302 |
| Pathogenic Escherichia coli infection-Homo sapiens (human) | 55 | 2 (3.6%) | 0.0070 | 0.0302 |
| Pathogenic Escherichia coli infection | 56 | 2 (3.6%) | 0.0072 | 0.0302 |
| Role of Calcineurin-dependent NFAT signaling in lymphocytes | 58 | 2 (3.4%) | 0.0077 | 0.0310 |
| Folate Metabolism | 66 | 2 (3.0%) | 0.0099 | 0.0381 |
| Cell Line | MRC-5 | HEK-293 | jFib | aFib |
|---|---|---|---|---|
| IC50 value (µM) | 2.4 ± 0.4 * | 9.4 ± 1.4 | 1.8 ± 0.8 | 6.9 ± 1.1 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kretschmer, N.; Deutsch, A.; Durchschein, C.; Rinner, B.; Stallinger, A.; Higareda-Almaraz, J.C.; Scheideler, M.; Lohberger, B.; Bauer, R. Comparative Gene Expression Analysis in WM164 Melanoma Cells Revealed That β-β-Dimethylacrylshikonin Leads to ROS Generation, Loss of Mitochondrial Membrane Potential, and Autophagy Induction. Molecules 2018, 23, 2823. https://doi.org/10.3390/molecules23112823
Kretschmer N, Deutsch A, Durchschein C, Rinner B, Stallinger A, Higareda-Almaraz JC, Scheideler M, Lohberger B, Bauer R. Comparative Gene Expression Analysis in WM164 Melanoma Cells Revealed That β-β-Dimethylacrylshikonin Leads to ROS Generation, Loss of Mitochondrial Membrane Potential, and Autophagy Induction. Molecules. 2018; 23(11):2823. https://doi.org/10.3390/molecules23112823
Chicago/Turabian StyleKretschmer, Nadine, Alexander Deutsch, Christin Durchschein, Beate Rinner, Alexander Stallinger, Juan Carlos Higareda-Almaraz, Marcel Scheideler, Birgit Lohberger, and Rudolf Bauer. 2018. "Comparative Gene Expression Analysis in WM164 Melanoma Cells Revealed That β-β-Dimethylacrylshikonin Leads to ROS Generation, Loss of Mitochondrial Membrane Potential, and Autophagy Induction" Molecules 23, no. 11: 2823. https://doi.org/10.3390/molecules23112823
APA StyleKretschmer, N., Deutsch, A., Durchschein, C., Rinner, B., Stallinger, A., Higareda-Almaraz, J. C., Scheideler, M., Lohberger, B., & Bauer, R. (2018). Comparative Gene Expression Analysis in WM164 Melanoma Cells Revealed That β-β-Dimethylacrylshikonin Leads to ROS Generation, Loss of Mitochondrial Membrane Potential, and Autophagy Induction. Molecules, 23(11), 2823. https://doi.org/10.3390/molecules23112823