Comparison and Identification for Rhizomes and Leaves of Paris yunnanensis Based on Fourier Transform Mid-Infrared Spectroscopy Combined with Chemometrics
Abstract
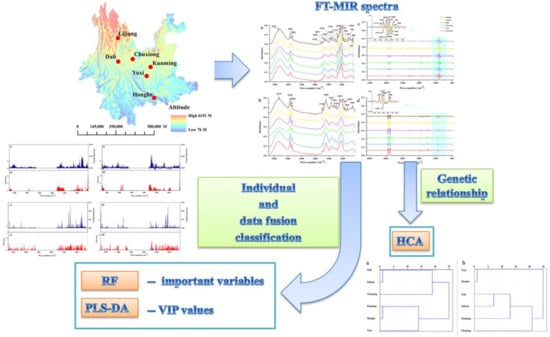
:1. Introduction
2. Results and Discussion
2.1. Comparison Analysis between Rhizomes and Leaves
2.2. Origin Traceability Based on Chemometrics
2.2.1. Using Rhizome FT-MIR Spectra Datasets
2.2.2. Using Leaf FT-MIR Spectra Datasets
2.3. Regional Differences between VIP and Important Variables
2.4. Data Fusion Strategy
2.5. Hierarchical Clustering Analysis
3. Materials and Methods
3.1. Plant Material Preparation
3.2. FT-MIR Spectral Acquisition
3.3. Chemometrics Methods
3.3.1. Principal Component Analysis
3.3.2. Partial Least Squares Discriminant Analysis
3.3.3. Random Forest
3.3.4. Hierarchical Cluster Analysis
3.4. Data Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Cunningham, A.B.; Brinckmann, J.A.; Bi, Y.F.; Pei, S.J.; Schippmann, U.; Luo, P. Paris in the spring: A review of the trade, conservation and opportunities in the shift from wild harvest to cultivation of Paris polyphylla (Trilliaceae). J. Ethnopharmacol. 2018, 222, 208–216. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Xu, J.H.; Chen, R.P.; Yang, H.; Liu, Y.P. Status of the genus Paris L. re-sources of Yunnan and countermeasures for protection. J. Yunnan Univ. 2006, 28, 307–310. [Google Scholar]
- State Pharmacopoeia Commission. Chinese Pharmacopoeia; Chemistry and Industry Press: Beijing, China, 2015. [Google Scholar]
- Li, H. The Genus Paris (Trilliaceae); Science Press: Beijing, China, 1998; pp. 12–16. [Google Scholar]
- Yang, J.; Wang, Y.H.; Li, H. Paris qiliangiana (Melanthiaceae), a new species from Hubei, China. Phytotaxa 2017, 329, 193–196. [Google Scholar] [CrossRef]
- Qin, X.J.; Yu, M.Y.; Ni, W.; Yan, H.; Chen, C.X.; Cheng, Y.C.; Li, H.; Liu, H.Y. Steroidal saponins from stems and leaves of Paris polyphylla var. yunnanensis. Phytochemistry 2016, 121, 20–29. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.W.; Feng, L.L.; Li, H.F. Analysis of differences and correlation of steroidal saponins in rhizomes and leaves of Paris polyphylla var. yunnanensis from different planting base. Chin. J. Exp. Tradit. Med. Form. 2018, 24, 41–48. [Google Scholar]
- Yang, Y.G.; Zhang, J.; Zhao, Y.L.; Zhang, J.Y.; Wang, Y.Z. Quantitative determination and evaluation of Paris polyphylla var. yunnanensis with different harvesting times using UPLC-UV-MS and FT-IR spectroscopy in combination with partial least squares discriminant analysis. Biomed. Chromatogr. 2017, 31. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.G.; Jin, H.; Zhang, J.; Zhang, J.Y.; Wang, Y.Z. Quantitative evaluation and discrimination of wild Paris polyphylla var. yunnanensis (Franch.) Hand.-Mazz from three regions of Yunnan Province using UHPLC-UV-MS and UV spectroscopy couple with partial least squares discriminant analysis. J. Nat. Med. 2017, 71, 148–157. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.Z.; Wen, F.Y.; Zhang, T.; Yang, Y.X.; Fang, Q.M.; Zhang, H.; Xue, D. Evaluation of saponins in Paris Polyphylla var. chinensis from twenty-one growing areas. Chin. Tradit. Patent Med. 2017, 39, 2345–2350. [Google Scholar]
- Wu, X.M.; Zhang, Q.Z.; Wang, Y.Z. Traceability of wild Paris polyphylla Smith var. yunnanensis based on data fusion strategy of FT-MIR and UV-Vis combined with SVM and random forest. Spectrochim. Acta A 2018, 205, 479–488. [Google Scholar] [CrossRef] [PubMed]
- Pei, Y.F.; Wu, L.H.; Zhang, Q.Z.; Wang, Y.Z. Geographical traceability of cultivated Paris polyphylla var. yunnanensis using ATR-FTMIR spectroscopy with three mathematical algorithms. Anal. Methods 2018. [Google Scholar] [CrossRef]
- Gad, H.A.; El-Ahmady, S.H.; Abou-Shoer, M.I.; Al-Azizi, M.M. Application of chemometrics in authentication of herbal medicines: A review. Phytochem. Anal. 2012, 24, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Biancolillo, A.; Marini, F. Chemometrics applied to plant spectral analysis. In Vibrational Spectroscopy for Plant Varieties and Cultivars Characterization; Elsevier: Amsterdam, The Netherlands, 2018; pp. 69–104. [Google Scholar]
- Li, J.; Zhang, J.; Zhao, Y.L.; Huang, H.Y.; Wang, Y.Z. Comprehensive quality assessment based specific chemical profiles for geographic and tissue variation in Gentiana rigescens using HPLC and FTIR method combined with principal component analysis. Front. Chem. 2017, 5. [Google Scholar] [CrossRef] [PubMed]
- Qi, L.M.; Liu, H.G.; Li, J.Q.; Li, T.; Wang, Y.Z. Feature fusion of ICP-AES, UV-Vis and FT-MIR for origin traceability of Boletus Edulis mushrooms in combination with chemometrics. Sensors 2018, 18, 241. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, J.Y.; Wang, Y.Z. FT-MIR and NIR spectral data fusion: A synergetic strategy for the geographical traceability of Panax notoginseng. Anal. Bioanal. Chem. 2018, 410, 91–103. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Huang, H.Y.; Zuo, Z.T.; Wang, Y.Z. Comprehensive quality assessment of Dendrubium officinale using ATR-FTIR spectroscopy combined with random forest and support vector machine regression. Spectrochim. Acta A 2018, 205, 637–648. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.G.; Wang, Y.Z. Characterization of Paris polyphylla var. yunnanensis by infrared and ultraviolet spectroscopies with chemometric data fusion. Anal. Lett. 2018, 51, 1730–1742. [Google Scholar] [CrossRef]
- Xie, J.D.; Sun, L. An overall quality evaluation of Paridis Rhizoma by multiple components determination based on the chemometrics. Chin. J. Pharm. Anal. 2015, 35, 1585–1590. [Google Scholar]
- Zhang, S.S.; Liu, X.; Wang, J.F.; Yu, M.J.; Huang, Z.J.; Liu, Y.; Zhang, H. Determination of seven steroidal saponins in Paridis Rhizoma and polygerm varieties from different regions in Yunnan Province by UPLC and establishment of fingerprint. Chin. Tradit. Herb. Drugs 2016, 47, 4257–4263. [Google Scholar]
- Zhang, J.Y.; Wang, Y.Z.; Zhao, Y.L.; Yang, S.B.; Zhang, J.; Yuan, T.J.; Wang, J.J.; Jin, H. Ultraviolet absorption spectrum analysis and identification of medicinal plants of Paris. Spectrosc. Spectr. Anal. 2012, 32, 2176–2180. [Google Scholar]
- Sun, S.Q.; Zhou, Q.; Chen, J.B. Analysis of Traditional Chinese Medicine by Infrared Spectroscopy; Chemical Industry Press: Beijing, China, 2010. [Google Scholar]
- Yang, L.F.; Ma, F.; Zhou, Q.; Sun, S.Q. Analysis and identification of wild and cultivated Paridis Rhizoma by infrared spectroscopy. J. Mol. Struct. 2018, 1165, 37–41. [Google Scholar] [CrossRef]
- Wold, S.; Esbensen, K.; Geladi, P. Principal component analysis. Chemometr. Intell. Lab. Syst. 1987, 2, 37–52. [Google Scholar] [CrossRef]
- Ringnér, M. What is principal component analysis? Nat. Biotechnol. 2008, 26, 303–304. [Google Scholar] [CrossRef] [PubMed]
- Jolliffe, I.T. Principal Component Analysis, 2nd ed.; Springer: New York, NY, USA, 2002. [Google Scholar]
- Ståle, L.; Wold, S. Partial least squares analysis with cross-validation for the two-class problem: A Monte Carlo study. J. Chemometr. 1987, 1, 185–196. [Google Scholar]
- Indahl, U.G.; Martens, H.; Naes, T. From dummy regression to prior probabilities in PLS-DA. J. Chemometr. 2007, 21, 529–536. [Google Scholar] [CrossRef]
- Nocairi, H.; Qannari, E.M.; Vigneau, E.; Bertrand, D. Discrimination on latent components with respect to patterns. Application to multicollinear data. Comput. Stat. Data Anal. 2005, 48, 139–147. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Amjad, A.; Ullah, R.; Khan, S.; Bilal, M.; Khan, A. Raman spectroscopy based analysis of milk using random forest classification. Vib. Spectrosc. 2018, 99, 124–129. [Google Scholar] [CrossRef]
- Saptoro, A.; Tadé, M.O.; Vuthaluru, H. A modified Kennard-Stone algorithm for optimal division of data for developing artificial neural network models. Chem. Prod. Process Model. 2012, 7, 1–14. [Google Scholar] [CrossRef]
- Rajer-Kanduč, K.; Zupan, J.; Majcen, N. Separation of data on the training and test set for modelling: A case study for modelling of five colour properties of a white pigment. Chemometr. Intell. Lab. Syst. 2003, 65, 221–229. [Google Scholar] [CrossRef]
- Jain, A.K.; Dubes, R.C. Algorithms for clustering data. In Technometrics; Prentice-Hall, Inc.: Englewood Cliffs, NJ, USA, 1988. [Google Scholar]
- Barnes, R.J.; Dhanoa, M.S.; Lister, S.J. Standard normal variate transformation and de-trending of near-infrared diffuse reflectance spectra. Appl. Spectrosc. 1989, 43, 772–777. [Google Scholar] [CrossRef]
- Savitzky, A.; Golay, M.J. Smoothing and differentiation of data by simplified least squares procedures. Anal. Chem. 1964, 36, 1627–1639. [Google Scholar] [CrossRef]
- Xie, L.J.; Ye, X.Q.; Liu, D.H.; Ying, Y.B. Quantification of glucose, fructose and sucrose in bayberry juice by NIR and PLS. Food Chem. 2009, 114, 1135–1140. [Google Scholar] [CrossRef]
- Qi, L.M.; Zhang, J.; Liu, H.G.; Li, T.; Wang, Y.Z. Fourier transform mid-infrared spectroscopy and chemometrics to identify and discriminate Boletus edulis and Boletus tomentipes mushrooms. Int. J. Food Prop. 2017, 20, S56–S68. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are not available from the authors. |









| Preprocessing | Set | Classes a | PLS-DA | RF | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| SENS | SPEC | ACC | MCC | SENS | SPEC | ACC | MCC | |||
| SNV-SD | Calibration set | 1 | 1 | 1 | 1 | 1 | 1 | 0.996 | 0.997 | 0.987 |
| 2 | 1 | 1 | 1 | 1 | 0.984 | 0.996 | 0.993 | 0.98 | ||
| 3 | 1 | 1 | 1 | 1 | 0.975 | 1 | 0.997 | 0.986 | ||
| 4 | 1 | 1 | 1 | 1 | 0.951 | 0.992 | 0.987 | 0.944 | ||
| 5 | 1 | 1 | 1 | 1 | 0.9831 | 1 | 0.997 | 0.989 | ||
| 6 | 1 | 1 | 1 | 1 | 1 | 0.996 | 0.997 | 0.99 | ||
| Validation set | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | ||
| 3 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | ||
| 4 | 1 | 1 | 1 | 1 | 0.95 | 1 | 0.994 | 0.971 | ||
| 5 | 1 | 1 | 1 | 1 | 1 | 0.992 | 0.994 | 0.979 | ||
| 6 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | ||
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pei, Y.-F.; Zhang, Q.-Z.; Zuo, Z.-T.; Wang, Y.-Z. Comparison and Identification for Rhizomes and Leaves of Paris yunnanensis Based on Fourier Transform Mid-Infrared Spectroscopy Combined with Chemometrics. Molecules 2018, 23, 3343. https://doi.org/10.3390/molecules23123343
Pei Y-F, Zhang Q-Z, Zuo Z-T, Wang Y-Z. Comparison and Identification for Rhizomes and Leaves of Paris yunnanensis Based on Fourier Transform Mid-Infrared Spectroscopy Combined with Chemometrics. Molecules. 2018; 23(12):3343. https://doi.org/10.3390/molecules23123343
Chicago/Turabian StylePei, Yi-Fei, Qing-Zhi Zhang, Zhi-Tian Zuo, and Yuan-Zhong Wang. 2018. "Comparison and Identification for Rhizomes and Leaves of Paris yunnanensis Based on Fourier Transform Mid-Infrared Spectroscopy Combined with Chemometrics" Molecules 23, no. 12: 3343. https://doi.org/10.3390/molecules23123343
APA StylePei, Y. -F., Zhang, Q. -Z., Zuo, Z. -T., & Wang, Y. -Z. (2018). Comparison and Identification for Rhizomes and Leaves of Paris yunnanensis Based on Fourier Transform Mid-Infrared Spectroscopy Combined with Chemometrics. Molecules, 23(12), 3343. https://doi.org/10.3390/molecules23123343