Quality Assessment of Kumu Injection, a Traditional Chinese Medicine Preparation, Using HPLC Combined with Chemometric Methods and Qualitative and Quantitative Analysis of Multiple Alkaloids by Single Marker
Abstract
:1. Introduction
2. Results and Discussion
2.1. The Establishment of Analytical Protocol
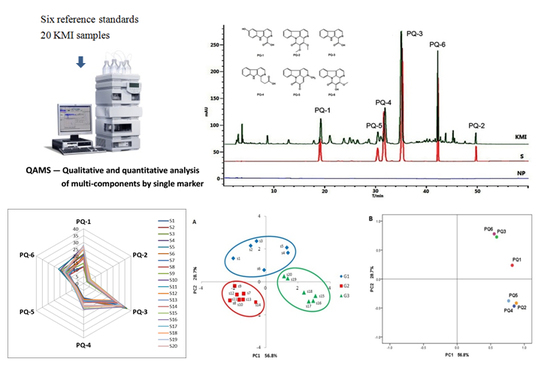
2.1.1. System Suitability and Chromatographic Peaks Identification
2.1.2. Method Validation
2.1.3. Assessment of the QAMS Method
2.2. Quality Evaluation of KMI Based on Six Components
2.2.1. Concentrations of the Six Components in KMI Samples
2.2.2. Hierarchical Cluster Analysis (HCA)
2.2.3. Radar Plot Analysis
2.2.4. Principal Component Analysis (PCA)
3. Materials and Methods
3.1. Materials and Reagents
3.2. Experimental Design
3.3. Preparation of Sample Solutions and Standard Solutions
3.4. HPLC System and Conditions
3.5. Determination of Relative Correction Factors (RCFs)
3.6. Data Analysis
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| ESM | External standard method |
| GAP | Good agriculture process |
| HCA | Hierarchical cluster analysis |
| HPLC | High performance liquid chromatography |
| KMI | Kumu injection |
| LOD | Limit of detection |
| LOQ | Limit of quantification |
| NP | Negative control preparation |
| PCA | Principal component analysis |
| QAMS | Qualitative and quantitative analysis of multi-component by single marker |
| RCF | Relative correction factor |
| RRT | Relative retention time |
| RPA | Relative peak area |
| RSD | Relative standard deviation |
| SD | Standard deviation |
| TCM | Traditional Chinese medicine |
References
- Hou, J.J.; Wu, W.Y.; Da, J.; Yao, S.; Long, H.L.; Yang, Z.; Guo, D.A. Ruggedness and robustness of conversion factors in method of simultaneous determination of multi-components with single reference standard. J. Chromatogr. A 2011, 1218, 5618–5627. [Google Scholar] [CrossRef] [PubMed]
- He, J.Y.; Zhu, S.; Goda, Y.; Cai, S.Q.; Komatsu, K. Quality evaluation of medicinally-used Codonopsis species and Codonopsis Radix based on the contents of pyrrolidine alkaloids, phenylpropanoid and polyacetylenes. J. Nat. Med. 2014, 68, 326–339. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.Q.; Jia, X.H.; Zhu, S.; Komatsu, K.; Wang, X.; Cai, S.Q. A systematic study on the influencing parameters and improvement of quantitative analysis of multi-component with single marker method using notoginseng as research subject. Talanta 2015, 134, 587–595. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Mao, F.; Zhao, Y.; Wang, W.; Fan, L.; Gao, X.; Zhao, J.; Tian, H. UPLC Quantitative Analysis of Multi-Components by Single Marker and Quality Evaluation of Polygala tenuifolia Wild. Extracts. Molecules 2017, 22, 2276. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Li, Q.; Liu, J.; Yin, Y.; Bi, K. Combinative method using multi-components quantitation by single reference standard and HPLC fingerprint for comprehensive evaluation of Rhodiola crenulata H. Ohba. Anal. Methods 2014, 6, 5891–5898. [Google Scholar] [CrossRef]
- Li, D.W.; Zhu, M.; Shao, Y.D.; Shen, Z.; Weng, C.C.; Yan, W.D. Determination and quality evaluation of green tea extracts through qualitative and quantitative analysis of multi-components by single marker (QAMS). Food Chem. 2016, 197, 1112–1120. [Google Scholar] [CrossRef] [PubMed]
- National Commission of Chinese Pharmacopoeia. Pharmacopoeia of People’s Republic of China; China Medical Science Press: Beijing, China, 2015; Volume I, p. 285. ISBN 978-7-5067-7337-9. [Google Scholar]
- National Commission of Chinese Pharmacopoeia. Prescription of Chinese Medicinal, Drug Standards Issued by Ministry of Public Health; China Medical Science Press: Beijing, China, 1998; p. 184. [Google Scholar]
- Zhao, W.; Chen, Z.W.; Sun, J.H.; He, Y.; Liu, W.; Hu, S.; Guang, Z.Y.; Chen, F.L. Alkaloid extraction from Picrasnia quassioides (D. Don) Benn. and safety of its compound injection. Guangxi Agric. Sci. 2012, 43, 865–868. [Google Scholar] [CrossRef]
- Chen, C.C.; Li, Z.R.; Liu, X.C. Observation on the curative effect of Kumu injection on acute upper respiratory infection in children. J. Chin. Prescr. Drug 2016, 14, 40–41. [Google Scholar] [CrossRef]
- Zhang, C.X. Clinical observation on treatment of diarrhea in children with acupoint injection of Kumu injection. Nei Mongol J. Tradit. Chin. Med. 2015, 6, 81. [Google Scholar] [CrossRef]
- Wu, W.H. Clinical observation on acute upper respiratory tract infection treated by Kumu injection. Chin. Community Dr. 2016, 22, 119–120. [Google Scholar] [CrossRef]
- Ishikura, M.; Abe, T.; Choshi, T.; Hibino, S. Simple indole alkaloids and those with a nonrearranged monoterpenoid unit. Nat. Prod. Rep. 2015, 32, 1389–1471. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.X.; Zhou, Y.J. Canthin-6-one alkaloids from Picrasma quassioides and their cytotoxic activity. J. Asian Nat. Prod. Res. 2008, 10, 1009–1012. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.F.; Shao, M.; Zhai, D.W.; Liu, K.; Wu, L.J. Protective effect of 4-methoxy-5-hydroxycanthin-6-one, a natural alkaloid, on dextran sulfate sodium induced rat colitis. Planta Med. 2009, 75, 142–145. [Google Scholar] [CrossRef] [PubMed]
- Fan, H.Y.; Qi, D.; Yang, M.Y.; Fang, H.; Liu, K.; Zhao, F. In vitro and in vivo anti-inflammatory effects of 4-methoxy-5-hydroxycanthin-6-one, a natural alkaloid from Picrasma quassioides. Phytomedicine 2013, 20, 319–323. [Google Scholar] [CrossRef] [PubMed]
- Jiao, W.H.; Gao, H.; Zhao, F.; Lin, H.W.; Pan, Y.M.; Zhou, G.X.; Yao, X.S. Anti-inflammatory alkaloids from the stems of Picrasma quassioides BENNET. Chem. Pharm. Bull. 2011, 59, 359–364. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.S.; Lee, Y.; Kwon, T.R.; Kim, Y.H.; Kim, B.J. Picrasma quassioides inhibits LPS- and IFN-γ-stimulated nitric oxide production and inflammatory response in RAW264.7 macrophage cells. Biotechnol. Bioprocess Eng. 2014, 19, 404–410. [Google Scholar] [CrossRef]
- Yang, W.H.; Xiao, S.X.; Zheng, J.H. Authentication of Kumu injection. Lishizhen Med. Mater. Med. Res. 2001, 12, 338. [Google Scholar]
- Huang, W.D.; Song, Y.X.; Lv, W.Q. Determination of canthinones alkaloids in Kumu injection by HPLC. Chin. J. Hosp. Pharm. 2010, 30, 1066–1067. [Google Scholar]
- Lai, Z.Q.; Ou, G.D.; Xiao, S.X.; Liao, Z.Y.; Liao, H.J.; Qin, Z.; Lin, J.; Su, Z.R. HPLC simultaneous determination of three active ingredients in Kumu injection. Chin. J. Pharm. Anal. 2011, 31, 15–18. [Google Scholar] [CrossRef]
- Lai, Z.Q.; Yi, Y.Y.; Liao, H.J.; Su, J.Y.; Liao, Z.Y.; Lin, J.; Su, Z.R. Fingerprint research and multi-component quantitative analysis of KMI by HPLC. Chin. J. Chin. Mater. Med. 2011, 36, 1739–1743. [Google Scholar] [CrossRef]
- Lai, Z.Q.; Liao, H.J.; Ip, S.P.; Yi, Y.Y.; Shi, S.J.; Su, J.Y.; Lai, X.P.; Su, Z.R.; Lin, Z.X. Liquid chromatography–mass spectrometry method for the simultaneous determination and confirmation of seven active components in Chinese medicine Kumu injection. Trop. J. Pharm. Res. 2014, 13, 141–148. [Google Scholar] [CrossRef]
- Ohmoto, T.; Koike, K. Studies on the constituents of Picrasma quassioides BENNET. III. Chem. Pharm. Bull. 1984, 32, 3579–3583. [Google Scholar] [CrossRef]
- Chen, M.; Fan, H.Y.; Dai, S.J.; Liu, K. Alkaloids from twigs and leaves of Picrasma quassioides. Chin. Tradit. Herb. Drugs 2007, 6, 807–810. [Google Scholar]
- Ohmoto, T.; Koike, K. Studies on the alkaloids from Picrasma quassioides BENNET. VI. Chem. Pharm. Bull. 1985, 33, 4901–4905. [Google Scholar] [CrossRef]
Sample Availability: Samples of the KMI and the compounds 6-hydroxy-β-carboline-1-carboxylic acid (PQ-1), 4,5-dimethoxycanthin-6-one (PQ-2), β-carboline-1-carboxylic acid (PQ-3), β-carboline-1-propanoic acid (PQ-4), 3-methylcanthin-5,6-dione (PQ-5), and nigakinone (PQ-6) are available from the authors. |





| Analytes | Stability | Precision | Repeatability | Reproducibility | Recovery | ||||
|---|---|---|---|---|---|---|---|---|---|
| (n = 5) | (n = 6) | (n = 6) | (n = 12) | (n = 6) | |||||
| RSD/% | RSD/% | RSD/% | RSD/% | RSD/% | |||||
| RRT | RPA | RRT | RPA | RRT | RPA | RRT | RPA | RPA | |
| PQ-1 | 0.7 | 3.1 | 1.7 | 1.4 | 2.2 | 5.5 | 3.2 | 6.0 | 0.6 |
| PQ-2 | 0.1 | 2.1 | 0.1 | 2.7 | 0.1 | 3.7 | 0.3 | 4.6 | 1.1 |
| PQ-3 | 0.2 | 0.6 | 0.8 | 3.4 | 1.0 | 5.4 | 2.7 | 5.2 | 1.4 |
| PQ-4 | 1.0 | 2.0 | 1.1 | 3.3 | 1.7 | 3.8 | 3.4 | 4.8 | 1.0 |
| PQ-5 | 0.6 | 4.4 | 0.9 | 3.8 | 1.4 | 2.6 | 2.8 | 5.4 | 0.7 |
| PQ-6 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.7 |
| Analytes | Regression Equation | R | LOD (μg·mL−1) | LOQ (μg·mL−1) | Linear Range(μg·mL−1) |
|---|---|---|---|---|---|
| PQ-1 | y = 42.681x − 59.049 | 0.9990 | 0.110 | 0.404 | 10.9–40.9 |
| PQ-2 | y = 65.376x − 5.5050 | 0.9997 | 0.066 | 0.241 | 2.0–9.8 |
| PQ-3 | y = 119.41x − 110.70 | 0.9998 | 0.039 | 0.139 | 20.6–62.0 |
| PQ-4 | y = 133.80x − 38.727 | 0.9997 | 0.033 | 0.122 | 5.2–31.3 |
| PQ-5 | y = 96.928x − 45.405 | 0.9991 | 0.058 | 0.188 | 1.5–14.1 |
| PQ-6 | y = 86.712x − 2.3510 | 0.9999 | 0.051 | 0.186 | 2.0–100.0 |
| Concentration Level | PQ-1 | PQ-2 | PQ-3 | PQ-4 | PQ-5 |
|---|---|---|---|---|---|
| C1 | 2.174 | 1.296 | 0.735 | 0.651 | 1.009 |
| C2 | 2.141 | 1.317 | 0.720 | 0.635 | 1.024 |
| C3 | 2.204 | 1.326 | 0.731 | 0.653 | 0.975 |
| C4 | 2.221 | 1.382 | 0.752 | 0.671 | 0.975 |
| C5 | 2.146 | 1.368 | 0.753 | 0.664 | 0.955 |
| C6 | 2.084 | 1.322 | 0.732 | 0.648 | 0.939 |
| Mean | 2.162 | 1.335 | 0.737 | 0.654 | 0.974 |
| RSD/% | 2.3 | 2.5 | 1.8 | 1.9 | 3.3 |
| Instrument | Column No. | fx/PQ-1 | fx/PQ-2 | fx/PQ-3 | fx/PQ-4 | fx/PQ-5 |
|---|---|---|---|---|---|---|
| Agilent 1100 | 1# | 2.162 | 1.335 | 0.737 | 0.654 | 0.974 |
| 2# | 2.182 | 1.353 | 0.745 | 0.663 | 0.990 | |
| 3# | 2.143 | 1.325 | 0.729 | 0.647 | 0.969 | |
| Shimadzu LC-20AT | 1# | 2.165 | 1.337 | 0.740 | 0.655 | 0.972 |
| 2# | 2.186 | 1.352 | 0.751 | 0.659 | 0.987 | |
| 3# | 2.146 | 1.325 | 0.730 | 0.649 | 0.967 | |
| Agilent 1260 | 1# | 2.160 | 1.340 | 0.740 | 0.655 | 0.980 |
| 2# | 2.185 | 1.357 | 0.746 | 0.660 | 1.001 | |
| 3# | 2.144 | 1.329 | 0.730 | 0.651 | 0.976 | |
| mean | 2.164 | 1.339 | 0.739 | 0.655 | 0.980 | |
| SD | 0.017 | 0.012 | 0.008 | 0.005 | 0.011 | |
| RSD/% | 0.8 | 0.9 | 1.1 | 0.8 | 1.2 |
| Sample No. | PQ-6 | PQ-1 | PQ-2 | PQ-3 | PQ-4 | PQ-5 | Total | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ESM | ESM | QAMS | SD | ESM | QAMS | SD | ESM | QAMS | SD | ESM | QAMS | SD | ESM | QAMS | SD | ||
| S1 | 15.54 | 19.45 | 19.46 | 0.01 | 2.77 | 2.77 | 0.00 | 32.81 | 32.82 | 0.01 | 10.51 | 10.50 | 0.01 | 5.07 | 5.08 | 0.01 | 86.18 |
| S2 | 17.67 | 21.30 | 21.31 | 0.01 | 2.95 | 2.95 | 0.00 | 36.25 | 36.27 | 0.01 | 11.19 | 11.18 | 0.01 | 5.42 | 5.44 | 0.01 | 94.82 |
| S3 | 18.93 | 21.74 | 21.74 | 0.01 | 2.97 | 2.97 | 0.00 | 36.70 | 36.72 | 0.01 | 11.51 | 11.50 | 0.01 | 5.55 | 5.57 | 0.01 | 97.44 |
| S4 | 19.61 | 27.19 | 27.20 | 0.01 | 3.91 | 3.91 | 0.00 | 35.95 | 35.97 | 0.01 | 12.51 | 12.50 | 0.01 | 7.05 | 7.07 | 0.01 | 106.27 |
| S5 | 21.31 | 24.51 | 24.52 | 0.01 | 4.00 | 4.00 | 0.00 | 35.87 | 35.89 | 0.01 | 12.41 | 12.40 | 0.01 | 6.97 | 6.98 | 0.01 | 105.11 |
| S6 | 16.38 | 18.92 | 18.90 | 0.02 | 3.73 | 3.73 | 0.00 | 35.02 | 35.00 | 0.01 | 12.41 | 12.44 | 0.02 | 6.80 | 6.78 | 0.01 | 93.22 |
| S7 | 14.62 | 12.49 | 12.47 | 0.01 | 3.27 | 3.27 | 0.00 | 29.20 | 29.18 | 0.01 | 13.12 | 13.15 | 0.02 | 6.93 | 6.92 | 0.01 | 79.60 |
| S8 | 12.15 | 12.19 | 12.21 | 0.01 | 2.98 | 2.98 | 0.00 | 28.21 | 28.19 | 0.01 | 12.36 | 12.36 | 0.00 | 6.59 | 6.59 | 0.00 | 74.48 |
| S9 | 15.71 | 14.75 | 14.76 | 0.00 | 2.79 | 2.79 | 0.00 | 26.47 | 26.48 | 0.01 | 12.19 | 12.18 | 0.01 | 6.37 | 6.38 | 0.01 | 78.30 |
| S10 | 11.93 | 21.11 | 21.08 | 0.02 | 3.61 | 3.61 | 0.00 | 26.22 | 26.20 | 0.01 | 12.39 | 12.41 | 0.02 | 5.90 | 5.89 | 0.01 | 81.12 |
| S11 | 12.67 | 19.77 | 19.74 | 0.02 | 3.75 | 3.75 | 0.00 | 25.58 | 25.56 | 0.01 | 12.38 | 12.40 | 0.02 | 5.94 | 5.92 | 0.01 | 80.04 |
| S12 | 14.00 | 19.43 | 19.41 | 0.02 | 3.67 | 3.66 | 0.00 | 24.60 | 24.58 | 0.01 | 11.85 | 11.87 | 0.02 | 5.83 | 5.81 | 0.01 | 79.34 |
| S13 | 13.95 | 17.89 | 17.86 | 0.02 | 3.89 | 3.89 | 0.00 | 25.31 | 25.30 | 0.01 | 12.83 | 12.86 | 0.02 | 6.18 | 6.17 | 0.01 | 80.02 |
| S14 | 13.76 | 18.16 | 18.19 | 0.02 | 4.34 | 4.34 | 0.00 | 27.65 | 27.64 | 0.01 | 14.07 | 14.07 | 0.00 | 6.48 | 6.48 | 0.00 | 84.47 |
| S15 | 16.83 | 25.06 | 25.09 | 0.02 | 6.04 | 6.04 | 0.00 | 34.53 | 34.51 | 0.01 | 18.49 | 18.50 | 0.00 | 7.58 | 7.58 | 0.00 | 108.56 |
| S16 | 15.83 | 25.23 | 25.26 | 0.02 | 5.73 | 5.73 | 0.00 | 34.24 | 34.22 | 0.01 | 18.96 | 18.96 | 0.00 | 7.71 | 7.71 | 0.00 | 107.73 |
| S17 | 15.90 | 27.85 | 27.86 | 0.01 | 5.26 | 5.27 | 0.00 | 31.36 | 31.38 | 0.01 | 17.56 | 17.55 | 0.01 | 8.00 | 8.02 | 0.01 | 105.98 |
| S18 | 16.98 | 25.17 | 25.18 | 0.01 | 5.64 | 5.65 | 0.00 | 32.62 | 32.63 | 0.01 | 17.08 | 17.07 | 0.01 | 7.29 | 7.31 | 0.01 | 104.81 |
| S19 | 17.19 | 26.89 | 26.90 | 0.01 | 4.47 | 4.48 | 0.00 | 32.22 | 32.24 | 0.01 | 16.33 | 16.32 | 0.01 | 6.39 | 6.40 | 0.01 | 103.53 |
| S20 | 16.82 | 27.63 | 27.65 | 0.01 | 4.40 | 4.41 | 0.00 | 32.24 | 32.26 | 0.01 | 15.85 | 15.84 | 0.01 | 6.12 | 6.14 | 0.01 | 103.10 |
| Mean | 15.89 | 21.34 | 4.01 | 31.15 | 13.80 | 6.51 | 92.71 | ||||||||||
| RSD/% | 15.4 | 22.7 | 24.9 | 13.3 | 18.7 | 12.1 | 13.2 | ||||||||||
| Sig. (2-tailed) | 0.53 | 0.36 | 0.78 | 0.61 | 0.19 | ||||||||||||
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, N.; Li, Z.-Y.; Zheng, X.-L.; Li, Q.; Yang, X.; Xu, H. Quality Assessment of Kumu Injection, a Traditional Chinese Medicine Preparation, Using HPLC Combined with Chemometric Methods and Qualitative and Quantitative Analysis of Multiple Alkaloids by Single Marker. Molecules 2018, 23, 856. https://doi.org/10.3390/molecules23040856
Wang N, Li Z-Y, Zheng X-L, Li Q, Yang X, Xu H. Quality Assessment of Kumu Injection, a Traditional Chinese Medicine Preparation, Using HPLC Combined with Chemometric Methods and Qualitative and Quantitative Analysis of Multiple Alkaloids by Single Marker. Molecules. 2018; 23(4):856. https://doi.org/10.3390/molecules23040856
Chicago/Turabian StyleWang, Ning, Zhi-Yong Li, Xiao-Li Zheng, Qiao Li, Xin Yang, and Hui Xu. 2018. "Quality Assessment of Kumu Injection, a Traditional Chinese Medicine Preparation, Using HPLC Combined with Chemometric Methods and Qualitative and Quantitative Analysis of Multiple Alkaloids by Single Marker" Molecules 23, no. 4: 856. https://doi.org/10.3390/molecules23040856
APA StyleWang, N., Li, Z. -Y., Zheng, X. -L., Li, Q., Yang, X., & Xu, H. (2018). Quality Assessment of Kumu Injection, a Traditional Chinese Medicine Preparation, Using HPLC Combined with Chemometric Methods and Qualitative and Quantitative Analysis of Multiple Alkaloids by Single Marker. Molecules, 23(4), 856. https://doi.org/10.3390/molecules23040856