Neo-5,22E-Cholestadienol Derivatives from Buthus martensii Karsch and Targeted Bactericidal Action Mechanisms
Abstract
:1. Introduction
2. Results and Discussion
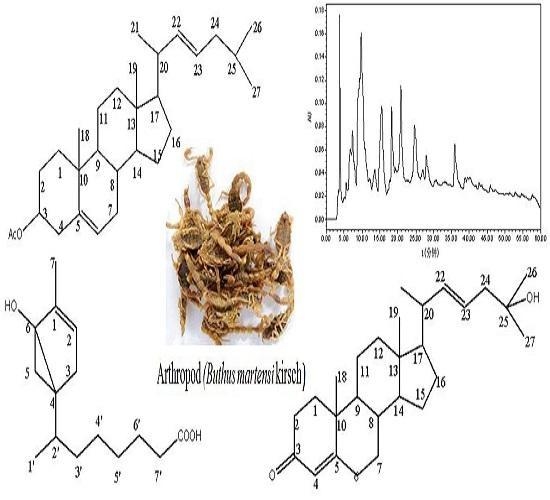
2.1. Chemistry
2.2. Assay of Antibacterial and Bactericidal Activities
3. Materials and Methods
3.1. General Experimental Procedure
3.2. Insect/Animal Materials
3.3. Extraction and Isolation
3.4. Determined Method of Antibacterial and the Bactericidal Activity
3.5. Molecular Docking Study
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Dossey, A.T. Insects and their chemical weaponry: New potential for drug discovery. Nat. Prod. Rep. 2010, 27, 1737–1757. [Google Scholar] [CrossRef] [PubMed]
- Matthew, G.; Schroeder, F.C. Comprehensive natural products II. Insect Natural Products; Elsevier: Oxford, UK, 2010; pp. 67–103. [Google Scholar]
- Simon, L.; Ruqaiyyah, S.; Naveed, A.K. Animals living in polluted environments are potential source of antimicrobials against infectious agents. Pathog. Glob. Health 2012, 106, 218–223. [Google Scholar]
- Niu, L.L.; Gao, J.Y.; Li, H.D.; Liu, J.N.; Yin, W.P. Novel skeleton compound Allomyrinanoid A and two purine alkaloids from the adult of Allomyrina dichotoma L. Bioorg. Med. Chem. Lett. 2016, 26, 366–369. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.W.; Liu, P.; Yin, W.P.; Jiang, Y.L.; Ren, Y.L. Isolation and identification of antibacterial neo-compounds from the red ants of ChangBai Mountain, Tetramorium sp. Bioorg. Med. Chem. Lett. 2012, 22, 2175–2181. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.Y.; Yin, W.P.; Gao, T.; Deng, R.X.; Li, X. Two bioactive compounds from the Chinese Scorpion Buthus martensi Karsch. Nat. Prod. Res. 2014, 28, 698–703. [Google Scholar] [CrossRef] [PubMed]
- Reich, H.J.; Jautelat, M.; Messe, M.T.; Weigert, F.J.; Roberts, J.D. 13C Nuclear magnetic resonance spectrum of gramicidin S-A, a decapeptide antibiotic. J. Am. Chem. Soc. 1969, 91, 7445–7454. [Google Scholar] [CrossRef]
- Mellado, G.G.; Zubã, A.E.; Ortega, M.J.; López-Gonzà lez, P.J. Steroids from the antarctic octocoral An-thomastus bathyproctus. J. Nat. Prod. 2005, 68, 1111–1115. [Google Scholar] [CrossRef]
- Echigo, S.; Castellanos, L.; Duque, C.; Uekusa, H.; Hara, N. C-24 stereochemistry of marine Sterols: (22E)-24-Ethyl-24-methylcholesta-5,22-dien-3β-ol and -24-Ethyl-24- methylcholest-5-en-3β-ol. J. Braz. Chem. Soc. 1991, 22, 997–1005. [Google Scholar] [CrossRef]
- Koreeda, M.; Harada, N.; Nakanishi, K. Exciton chirality method as applied to conjugated enones, esters, and lactones. J. Am. Chem. Soc. 1974, 96, 266–268. [Google Scholar] [CrossRef]
- CAS Registry Number: 1089664-70-9, (3β-acetate, 20S, 22E)-Cholesta-5,22-dien-3-ol. in SciFinder. Available online: https://m.chemsrc.com/en/cas/23515-91-5_943583.html (accessed on 4 November 2018).
- Raharivelomanana, P.; Robert, F.; Cambon, A.; Azzaro, M. β-Acoradienol, a sesquiterpene alcohol from Neocallitropsis pancheri. J. Nat. Prod. 1992, 55, 235–236. [Google Scholar] [CrossRef]
- Herz, W.; Sharma, P. Pycnolide, a seco-germacradienolide from Liatris pycnostachya, and other antitumor constituents of Liatris species. J. Org. Chem. 1976, 41, 1248–1253. [Google Scholar] [CrossRef] [PubMed]
- CAS Registry Number: (1) 87978-39-0; (2) 123887-29-6; in SciFinder. Available online: https://m.chemsrc.com/en/cas/7779-73-9_1118571.html (accessed on 4 November 2018).
- Ferraro, M.J.; Craig, W.A.; Dudley, M.N.G.; Eliopoulos, M.; Hecht, D.W.; Handler, J.; Roller, L.B.; Sheldon, A.T.; Swenson, J.M.; Ten over, F.C.; et al. Performance Standards for Antimicrobial Susceptibility Testing; 11th Informational Supplement; National Committee for Clinical Laboratory Standards: Wayne, PA, USA, 2000. [Google Scholar]
- Ayala-Núñez, N.V.; Villegas, H.H.L.; Turrent, L.d.C.I.; Padilla, C.R. Silver nanoparticles toxicity and bactericidal effect against methicillin-resistant Staphylococcus aureus: Nanoscale does matter. NanoBiotechnology 2009, 5, 1–9. [Google Scholar] [CrossRef]
- Konaté, K.; Mavoungou, J.F.; Lepengué, A.N.; Samseny, R.R.R.A.; Hilou, A.; Souza, A.; Dicko, M.H.; M’Batchi, B. Antibacterial activity against β-lactamase producing Methicillin and Ampicillin-resistants Staphylococcus aureus: Fractional inhibitory concentration index (FICI) determination. Ann. Clin. Microbiol. Antimicrob. 2012, 11. [Google Scholar] [CrossRef] [PubMed]
- Clinical and Laboratory Standards Institute. Methods for Determining Bactericidal Activity of Antimicrobial Agents; Approved Guideline; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 1999. [Google Scholar]
- May, J.; Shannon, K.; King, A.; French, G. Glycopeptide tolerance in Staphylococcus aureus. J. Antimicrob. Chemother. 1998, 42, 189–197. [Google Scholar] [CrossRef] [PubMed]
- Maria, M.; Traczewski, B.D.; Katz, J.N.S.; Steven, D.B. Inhibitory and bactericidal activities of Daptomycin, Vancomycin, and Teicoplanin against Methicillin-Resistant Staphylococcus aureus isolates collected from 1985 to 2007. Antimicrob. Agents Chemother. 2009, 53, 1735–1738. [Google Scholar]
- Rouse, M.S.; Steckelberg, J.M.; Patel, R. In vitro activity of ceftobiprole, daptomycin, linezolid, and vancomycin against methicillin-resistant staphylococci associated with endocarditis and bone and joint infection. Diagn. Microbiol. Infect. Dis. 2007, 58, 363–365. [Google Scholar] [CrossRef] [PubMed]
- Nicholas, S.B.; Nimish, P.; Theresa, I.S.; Wissam, I.E.; Atrouni, R.T.H.; Molly, E.S. Relationship between vancomycin tolerance and clinical outcomes in Staphylococcus aureus bacteraemia. J. Antimicrob. Chemother. 2017, 72, 535–542. [Google Scholar]
- Biedenbach, D.J.; Bell, J.M.; Sader, H.S.; Fritsche, T.R.; Jones, R.N.; Turnidge, J.D. Antimicrobial. susceptibility of Gram-positive bacterial isolates from the Asia-Pacific region and an in vitro evaluation of the bactericidal activity of daptomycin, vancomycin, and teicoplanin: A SENTRY Program Report (2003–2004). Int. J. Antimicrob. Agents 2007, 30, 143–149. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Ouyang, S.; Yu, B.; Liu, Y.; Huang, K.; Gong, J.; Zheng, S.; Li, Z.; Li, H.; Jiang, H. PharmMapper server. Nucleic Acids Res. 2010, 38, W609–W614. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Yuan, Z.; Huang, W.X.; Zheng, X.P. A clinical trial of doxycyclinc hydrochloride for injection in the treatment of acute bacterial infections. Chin. J. Antibiot. 2006, 31, 675–678. (In Chinese) [Google Scholar]
- Abouelfetouh, A.; Kassem, M.; Naguib, M.; El-Nakeeb, M. Investigation and treatment of fusidic acid resistance among Methicillin-Resistant Staphylococcal isolates from Egypt. Micro. Drug Resis. 2017, 23, 8–17. [Google Scholar] [CrossRef] [PubMed]
- Farrell, D.J.; Mendes, R.E.; Castanheira, M.; Jones, R.N. Activity of Fusidic acid tested against Staphylococci isolated from Patients in US Medical Centers in 2014. Antimicrob. Agents Chemother. 2016, 60, 3827–3831. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds 1, 2 and 3 are available from the authors. |














| No. | Compound 1 | Compound 2 | ||
|---|---|---|---|---|
| δH (J, Hz) | δc (DEPT) | δH (J, Hz) | δc (DEPT) | |
| 1 | 1.57 m, 1.20 m | 38.3 (CH2) | 1.02 m, 2.32 m | 36.2 (CH2) |
| 2 | 2.24 (2H, m) | 35.7 (CH2) | 1.86 m, 1.55 m | 27.2 (CH2) |
| 3 | - | 202.3 (C) | 3.50 m | 71.8 (CH) |
| 4 | 5.69 (1H, d, 1.48) | 126.1 (CH) | 1.82 m | 33.1 (CH2) |
| 5 | - | 165.1 (C) | 140.7 (C) | |
| 6 | 2.39 m | 38.7 (CH2) | 5.33 (1H, dd, 3.56, 1.52) | 121.7 (CH) |
| 7 | 1.02 m, 1.93 m | 31.2 (CH2) | 2.27 m, 1.25 m | 31.4 (CH2) |
| 8 | 1.60 m | 29.7 (CH) | 1.56 m | 31.9 (CH) |
| 9 | 0.91 m | 45.4 (CH) | 0.90 m | 50.1 (CH) |
| 10 | - | 42.3 (C) | - | 36.2 (C) |
| 11 | 1.50 m, 1.33 m | 21.2 (CH2) | 1.50 m, 1.06 m | 21.1 (CH2) |
| 12 | 1.18 m, 1.96 m | 39.5 (CH2) | 1.30 m, 2.30 m | 39.7 (CH2) |
| 13 | - | 50.0 (CH) | - | 56.1 (CH) |
| 14 | 0.90 m | 42.8 (CH) | 2.33 (1H, m) | 42.3 (CH) |
| 15 | 1.26 m, 2.40 m | 23.8 (CH2) | 1.30 m, 2.30 m | 24.7 (CH2) |
| 16 | 1.36 m, 2.02 m | 28.1 (CH2) | 1.86 m, 1.49 m | 28.5 (CH2) |
| 17 | 1.58 (1H, dd, 3.78, 1.36) | 54.8 (CH) | 1.16 m | 56.7 (CH) |
| 18 | 0.84 s | 12.0 (CH3) | 0.84 s | 11.9 (CH3) |
| 19 | 0.78 s | 17.3 (CH3) | 0.67 s | 18.7 (CH3) |
| 20 | 1.18 (1H, dd, 3.78, 1.48) | 36.3 (CH) | 1.06 m | 37.3 (CH) |
| 21 | 0.86 s | 18.9 (CH3) | 0.97 s | 18.9 (CH3) |
| 22 | 5.68 (1H, dd, 3.24, 1.48) | 131.9 (CH) | 5.35 (1H, dd, 3.56, 1.64) | 131.7 (CH) |
| 23 | 5.20 (1H, dd, 7.60, 3.28) | 135.6 (CH) | 5.16 (1H, dd, 6.68, 3.56) | 135.8 (CH) |
| 24 | 1.33 m, 2.40 m | 39.5 (CH2) | 1.12 m, 2.32 m | 29.7 (CH2) |
| 25 | 3.67 (1H, m) | 70.5 (C) | 2.27 (1H, m) | 28.5 (CH) |
| 26 | 0.87 s | 22.7 (CH3) | 1.03 (3H, d, 6.9) | 22.5 (CH3) |
| 27 | 0.90 s | 22.8 (CH3) | 1.06 (3H, d, 6.9) | 22.8 (CH3) |
| -OCO | - | - | 179.0 (C) | |
| -CH3 | - | 1.08 (s) | 28.0 (CH3) | |
| No. | δH (J, Hz) | δc (DEPT) | HMBC |
|---|---|---|---|
| 1 | 130.0 (C) | ||
| 2 | 5.34 (1H, br. s) | 129.7 (CH) | H-2/C-3, C-1 |
| 3 | 2.01 (2H, br. s) | 27.2 (CH2) | H-3/C-2, C-2′, |
| 4 | 29.3 (C) | ||
| 5 | 1.62 (2H, br. s) | 24.9 (CH2) | |
| 6 | 77.2 (C) | H-7/C-6 | |
| 7 | 0.87 (3H, s) | 14.0 (CH3) | H-7/C-3, C-2 |
| 1′ | 0.88 (3H, s) | 14.1 (CH3) | |
| 2′ | 1.26 (1H, br. s) | 29.1 (CH) | H-2′/C-4 |
| 3′ | 2.30 (2H, br. s) | 34.1 (CH2) | H-3′/C-5, C-4′ |
| 4′ | 1.26 (2H, br. s) | 32.0 (CH2) | H-4′/C-1′ |
| 5′ | 1.26 (2H, br. s) | 29.5 (CH2) | |
| 6′ | 1.26 (2H, br. s) | 29.7 (CH2) | |
| 7′ | 1.26 (2H, br. s) | 22.7 (CH2) | H-7′/C-5′, C-4′ |
| -COOH | 7.50 (1H) | 179.1 (C) |
| Strains | Compound 1 | Compound 2 | Compound 3 | Countrol | |||||
|---|---|---|---|---|---|---|---|---|---|
| MIC | MBC | MIC | MBC | MIC | MBC | Penicillin G | Gentamycin | Vancomycin | |
| B. subtilis (ATCC 6051) | 78 | >128 | 78 | >128 | 256 | >256 | - | - | - |
| S. aureus (ATCC 6538) | 78 | >128 | 16 | 32 | >256 | >256 | - | - | - |
| E. coli (ATCC 25922) | 256 | >256 | 256 | >256 | 256 | >256 | - | 6 | - |
| P. aeruginosa (ATCC 27853) | 64 | >78 | 16 | <48 | >256 | >256 | - | 6 | - |
| MRSA (Clinical isolated) | 256 | >256 | 128 | 256 | >256 | >256 | - | - | 0.5 |
| Ligand Name | Absolute Energy | Conf Number | Relative Energy | LibDock Score |
|---|---|---|---|---|
| Doxycycline | 76.5451 | 2 | 2.29878 | 99.0843 |
| Compound 1 | 50.3676 | 38 | 10.4028 | 94.1788 |
| Compound 2 | 54.5212 | 55 | 9.67329 | 98.2142 |
| Ligand Name | Absolute Energy | Conf Number | Relative Energy | LibDock Score |
|---|---|---|---|---|
| Fusidic acid | 92.4881 | 51 | 13.1839 | 122.250 |
| Compound 1 | 60.4528 | 81 | 15.6049 | 124.669 |
| Compound 2 | 47.9485 | 22 | 7.98374 | 125.637 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lv, B.; Yin, W.; Gao, J.; Liu, H.; Liu, K.; Bai, J.; Yang, Q. Neo-5,22E-Cholestadienol Derivatives from Buthus martensii Karsch and Targeted Bactericidal Action Mechanisms. Molecules 2019, 24, 72. https://doi.org/10.3390/molecules24010072
Lv B, Yin W, Gao J, Liu H, Liu K, Bai J, Yang Q. Neo-5,22E-Cholestadienol Derivatives from Buthus martensii Karsch and Targeted Bactericidal Action Mechanisms. Molecules. 2019; 24(1):72. https://doi.org/10.3390/molecules24010072
Chicago/Turabian StyleLv, Biyu, Weiping Yin, Jiayu Gao, Huaqing Liu, Kun Liu, Jie Bai, and Qiangqiang Yang. 2019. "Neo-5,22E-Cholestadienol Derivatives from Buthus martensii Karsch and Targeted Bactericidal Action Mechanisms" Molecules 24, no. 1: 72. https://doi.org/10.3390/molecules24010072
APA StyleLv, B., Yin, W., Gao, J., Liu, H., Liu, K., Bai, J., & Yang, Q. (2019). Neo-5,22E-Cholestadienol Derivatives from Buthus martensii Karsch and Targeted Bactericidal Action Mechanisms. Molecules, 24(1), 72. https://doi.org/10.3390/molecules24010072