Bulged and Canonical G-Quadruplex Conformations Determine NDPK Binding Specificity
Abstract
:1. Introduction
2. Results
2.1. hex4_A5U Adopts a G4 Conformation in the Presence of Cations
2.2. hex4_A5U Oligonucleotide is a Mix of G4 Conformations
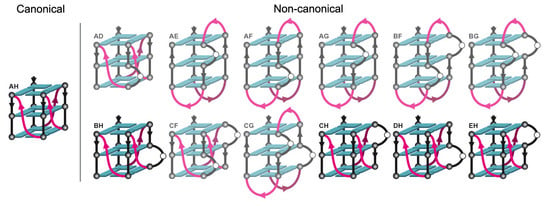
2.3. Locked hex4_A5U Variants Form G4s with Distinct Properties
2.4. ZmNDPK1 Requires Two Consecutive G-Tracts with a Single One-Base Loop for Efficient Binding
2.5. ZmNDPK1 Binds Intermolecular and Intramolecular G4s
2.6. ZmNDPK1 and Trim_A5U form a Heterogeneous Protein: Nucleic Acid Complex
3. Discussion
3.1. G4s in the Stress Response
3.2. Limitations in G4 Characterization Affect Analysis of hex4_A5U
3.3. hex4_A5U and its Truncated Variant trim_A5U are Highly Polymorphic G4-Forming Sequences
3.4. The G4-Binding Protein ZmNDPK1 Recognizes a Subset of Conformations Adopted by hex4_A5U DNA and Forms Filamentous Structures upon Binding
3.5. Generalization of G4 Heterogeneity across Domains of Life
4. Materials and Methods
4.1. Oligonucleotide and Protein Preparation
4.2. Absorption Spectrophotometry
4.3. Circular Dichroism Spectrophotometry
4.4. Dimethyl Sulfate (DMS) Footprinting
4.5. Nitrocellulose Filter Binding Assays for ZmNDPK1/G4 DNA Binding Affinity Analysis
4.6. Analytical Ultracentrifugation (AUC)
4.7. Fluorescence Resonance Energy Transfer (FRET)
4.8. Electron Microscopy (EM)
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Watson, J.D.; Crick, F.H. Genetical implications of the structure of deoxyribonucleic acid. Nature 1953, 171, 964–967. [Google Scholar] [CrossRef]
- Gellert, M.; Lipsett, M.N.; Davies, D.R. Helix formation by guanylic acid. Proc. Natl. Acad. Sci. USA 1962, 48, 2013–2018. [Google Scholar] [PubMed]
- Zimmerman, S.B.; Cohen, G.H.; Davies, D.R. X-ray fiber diffraction and model-building study of polyguanylic acid and polyinosinic acid. J. Mol. Biol. 1975, 92, 181–192. [Google Scholar] [PubMed]
- Harris, L.M.; Merrick, C.J. G-quadruplexes in pathogens: A common route to virulence control? PLoS Pathog. 2015, 11, e1004562. [Google Scholar] [CrossRef]
- Biffi, G.; Tannahill, D.; McCafferty, J.; Balasubramanian, S. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 2013, 5, 182–186. [Google Scholar] [CrossRef] [PubMed]
- Farhath, M.M.; Thompson, M.; Ray, S.; Sewell, A.; Balci, H.; Basu, S. G-Quadruplex-Enabling Sequence within the Human Tyrosine Hydroxylase Promoter Differentially Regulates Transcription. Biochemistry 2015, 54, 5533–5545. [Google Scholar] [PubMed]
- Lemmens, B.; van Schendel, R.; Tijsterman, M. Mutagenic consequences of a single G-quadruplex demonstrate mitotic inheritance of DNA replication fork barriers. Nat. Commun. 2015, 6, 8909. [Google Scholar]
- Kanoh, Y.; Matsumoto, S.; Fukatsu, R.; Kakusho, N.; Kono, N.; Renard-Guillet, C.; Masuda, K.; Iida, K.; Nagasawa, K.; Shirahige, K.; et al. Rif1 binds to G quadruplexes and suppresses replication over long distances. Nat. Struct. Mol. Biol. 2015, 22, 889–897. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Shin-ya, K.; Brosh, R.M. FANCJ Helicase Defective in Fanconia Anemia and Breast Cancer Unwinds G-Quadruplex DNA To Defend Genomic Stability. Mol. Cell. Biol. 2008, 28, 4116–4128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paeschke, K.; Capra, J.A.; Zakian, V.A. DNA Replication through G-Quadruplex Motifs Is Promoted by the Saccharomyces cerevisiae Pif1 DNA Helicase. Cell 2011, 145, 678–691. [Google Scholar] [Green Version]
- Smestad, J.A.; Maher, L.J. Relationships between putative G-quadruplex-forming sequences, RecQ helicases, and transcription. BMC Med. Genet. 2015, 16, 91. [Google Scholar] [CrossRef] [PubMed]
- Bartas, M.; Čutová, M.; Brázda, V.; Kaura, P.; Šťastný, J.; Kolomazník, J.; Coufal, J.; Goswami, P.; Červeň, J.; Pečinka, P. The Presence and Localization of G-Quadruplex Forming Sequences in the Domain of Bacteria. Molecules 2019, 24, 1711. [Google Scholar] [CrossRef] [PubMed]
- Ruggiero, E.; Tassinari, M.; Perrone, R.; Nadai, M.; Richter, S.N. The Long Terminal Repeat promoter of Retroviruses contains stable and conserved G-quadruplexes. ACS Infect. Dis. 2019. [Google Scholar] [CrossRef]
- Scalabrin, M.; Frasson, I.; Ruggiero, E.; Perrone, R.; Tosoni, E.; Lago, S.; Tassinari, M.; Palù, G.; Richter, S.N. The cellular protein hnRNP A2/B1 enhances HIV-1 transcription by unfolding LTR promoter G-quadruplexes. Sci. Rep. 2017, 7, 45244. [Google Scholar] [CrossRef] [PubMed]
- Eddy, J.; Maizels, N. Gene function correlates with potential for G4 DNA formation in the human genome. Nucleic Acids Res. 2006, 34, 3887–3896. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andorf, C.M.; Kopylov, M.; Dobbs, D.; Koch, K.E.; Stroupe, M.E.; Lawrence, C.J.; Bass, H.W. G-quadruplex (G4) motifs in the maize (Zea mays L.) genome are enriched at specific locations in thousands of genes coupled to energy status, hypoxia, low sugar, and nutrient deprivation. J. Genet. Genom. 2014, 41, 627–647. [Google Scholar] [CrossRef]
- Kalra, P.; Mishra, S.K.; Kaur, S.; Kumar, A.; Prasad, H.K.; Sharma, T.K.; Tyagi, J.S. G-Quadruplex-Forming DNA Aptamers Inhibit the DNA-Binding Function of HupB and Mycobacterium tuberculosis Entry into Host Cells. Mol. Ther. Nucleic Acids 2018, 13, 99–109. [Google Scholar] [CrossRef] [Green Version]
- Verma, A.; Yadav, V.K.; Basundra, R.; Kumar, A.; Chowdhury, S. Evidence of genome-wide G4 DNA-mediated gene expression in human cancer cells. Nucleic Acids Res 2009, 37, 4194–4204. [Google Scholar] [CrossRef] [Green Version]
- Sengupta, A.; Ganguly, A.; Chowdhury, S. Promise of G-Quadruplex Structure Binding Ligands as Epigenetic Modifiers with Anti-Cancer Effects. Molecules 2019, 24, 582. [Google Scholar] [CrossRef]
- Métifiot, M.; Amrane, S.; Litvak, S.; Andreola, M.L. G-quadruplexes in viruses: Function and potential therapeutic applications. Nucleic Acids Res. 2014, 42, 12352–12366. [Google Scholar]
- Ruggiero, E.; Richter, S.N. G-quadruplexes and G-quadruplex ligands: Targets and tools in antiviral therapy. Nucleic Acids Res. 2018, 46, 3270–3283. [Google Scholar] [CrossRef]
- Saranathan, N.; Vivekanandan, P. G-Quadruplexes: More Than Just a Kink in Microbial Genomes. Trends Microbiol. 2019, 27, 148–163. [Google Scholar] [CrossRef] [PubMed]
- Amrane, S.; Kerkour, A.; Bedrat, A.; Vialet, B.; Andreola, M.L.; Mergny, J.L. Topology of a DNA G-quadruplex structure formed in the HIV-1 promoter: A potential target for anti-HIV drug development. J. Am. Chem. Soc. 2014, 136, 5249–5252. [Google Scholar] [CrossRef]
- Hudson, J.S.; Ding, L.; Le, V.; Lewis, E.; Graves, D. Recognition and binding of human telomeric G-quadruplex DNA by unfolding protein 1. Biochemistry 2014, 53, 3347–3356. [Google Scholar] [CrossRef]
- Johnson, J.E.; Cao, K.; Ryvkin, P.; Wang, L.S.; Johnson, F.B. Altered gene expression in the Werner and Bloom syndromes is associated with sequences having G-quadruplex forming potential. Nucleic Acids Res. 2010, 38, 1114–1122. [Google Scholar] [CrossRef] [PubMed]
- Khateb, S.; Weisman-Shomer, P.; Hershco, I.; Loeb, L.A.; Fry, M. Destabilization of tetraplex structures of the fragile X repeat sequence (CGG)n is mediated by homolog-conserved domains in three members of the hnRNP family. Nucleic Acids Res. 2004, 32, 4145–4154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- London, T.B.C.; Barber, L.J.; Mosedale, G.; Kelly, G.P.; Balasubramanian, S.; Hickson, I.D.; Boulton, S.J.; Hiom, K. FANCJ is a structure-specific DNA helicase associated with the maintenance of genomic G/C tracts. J. Biol. Chem. 2008, 283, 36132–36139. [Google Scholar] [CrossRef] [PubMed]
- Qureshi, M.H.; Ray, S.; Sewell, A.L.; Basu, S.; Balci, H. Replication protein A unfolds G-quadruplex structures with varying degrees of efficiency. J. Phys. Chem. B 2012, 116, 5588–5594. [Google Scholar] [CrossRef] [PubMed]
- Safa, L.; Delagoutte, E.; Petruseva, I.; Alberti, P.; Lavrik, O.; Riou, J.-F.; Saintomé, C. Binding polarity of RPA to telomeric sequences and influence of G-quadruplex stability. Biochimie 2014, 103, 80–88. [Google Scholar] [CrossRef]
- Dempsey, L.A.; Sun, H.; Hanakahi, L.A.; Maizels, N. G4 DNA binding by LR1 and its subunits, nucleolin and hnRNP D, A role for G-G pairing in immunoglobulin switch recombination. J. Biol. Chem. 1999, 274, 1066–1071. [Google Scholar] [CrossRef]
- Hanakahi, L.A.; Sun, H.; Maizels, N. High affinity interactions of nucleolin with G-G-paired rDNA. J. Biol. Chem. 1999, 274, 15908–15912. [Google Scholar] [CrossRef]
- Quante, T.; Otto, B.; Brázdová, M.; Kejnovská, I.; Deppert, W.; Tolstonog, G.V. Mutant p53 is a transcriptional co-factor that binds to G-rich regulatory regions of active genes and generates transcriptional plasticity. Cell Cycle 2012, 11, 3290–3303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kopylov, M.; Bass, H.W.; Stroupe, M.E. The maize (Zea mays L.) nucleoside diphosphate kinase1 (ZmNDPK1) gene encodes a human NM23-H2 homologue that binds and stabilizes G-quadruplex DNA. Biochemistry 2015, 54, 1743–1757. [Google Scholar] [CrossRef]
- Webba da Silva, M. Geometric formalism for DNA quadruplex folding. Chemistry 2007, 13, 9738–9745. [Google Scholar] [CrossRef] [PubMed]
- Mukundan, V.T.; Phan, A.T. Bulges in G-quadruplexes: Broadening the definition of G-quadruplex-forming sequences. J. Am. Chem. Soc. 2013, 135, 5017–5028. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Correia, J.J.; Wang, L.; Trent, J.O.; Chaires, J.B. Not so crystal clear: The structure of the human telomere G-quadruplex in solution differs from that present in a crystal. Nucleic Acids Res. 2005, 33, 4649–4659. [Google Scholar] [CrossRef] [PubMed]
- Dailey, M.M.; Miller, M.C.; Bates, P.J.; Lane, A.N.; Trent, J.O. Resolution and characterization of the structural polymorphism of a single quadruplex-forming sequence. Nucleic Acids Res. 2010, 38, 4877–4888. [Google Scholar] [CrossRef] [PubMed]
- Seenisamy, J.; Rezler, E.M.; Powell, T.J.; Tye, D.; Gokhale, V.; Joshi, C.S.; Siddiqui-Jain, A.; Hurley, L.H. The dynamic character of the G-quadruplex element in the c-MYC promoter and modification by TMPyP4. J. Am. Chem Soc. 2004, 126, 8702–8709. [Google Scholar] [CrossRef]
- Miannay, F.; Banyasz, A.; Gustavsson, T.; Markovitsi, S. Excited States and Energy Transfer in G-Quadruplexes. J. Phys. Chem. C 2009, 113, 11760–11765. [Google Scholar] [CrossRef] [Green Version]
- Randazzo, A.; Spada, G.P.; da Silva, M.W. Circular dichroism of quadruplex structures. Top. Currr. Chem. 2013, 330, 67–86. [Google Scholar]
- Sun, D.; Jurley, L.H. Biochemical techniques for the characterization of G-quadruplex structures: EMSA, DMS footprinting, and DNA polymerase stop assay. Methods Mol. Bio. 2010, 608, 65–79. [Google Scholar]
- Harkness, R.W.t.; Mittermaier, A.K. G-register exchange dynamics in guanine quadruplexes. Nucleic Acids Res. 2016, 44, 3481–3494. [Google Scholar] [CrossRef]
- Mergny, J.-L.; Lacroix, L. Analysis of thermal melting curves. Oligonucleotides 2003, 13, 515–537. [Google Scholar] [CrossRef] [PubMed]
- Mergny, J.L.; Phan, A.T.; Lacroix, L. Following G-quartet formation by UV-spectroscopy. FEBS Lett. 1998, 435, 74–78. [Google Scholar] [CrossRef] [Green Version]
- Uzman, A. Molecular Cell Biology (4th edition) Harvey Lodish, Arnold Berk, S. Lawrence Zipursky, Paul Matsudaira, David Baltimore and James Darnell; Freeman & Co., New York, NY, 2000, 1084 pp., list price $102.25, ISBN 0-7167-3136-3. Biochem. Mol. Biol. Educ. 2001, 29, 126–128. [Google Scholar]
- Balagurumoorthy, P.; Brahmachari, S.K.; Mohanty, D.; Bansal, M.; Sasisekharan, V. Hairpin and parallel quartet structures for telomeric sequences. Nucleic Acids Res. 1992, 20, 4061–4067. [Google Scholar] [CrossRef]
- Cao, W.; Demeler, B. Modeling analytical ultracentrifugation experiments with an adaptive space-time finite element solution for multicomponent reacting systems. Biophys. J. 2008, 95, 54–65. [Google Scholar] [CrossRef]
- Demeler, B. UltraScan—A Comprehensive Data Analysis Software Package for Analytical Ultracentrifugation Experiments; Scott, D.J., Harding, S.E., Rowe, A.J., Eds.; Royal Society of Chemistry: Cambridge, UK, 2005; pp. 210–230. [Google Scholar]
- Huppert, J.L.; Balasubramanian, S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005, 33, 2908–2916. [Google Scholar] [CrossRef] [Green Version]
- Rhodes, D.; Lipps, H.J. G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 2015, 43, 8627–8637. [Google Scholar] [CrossRef] [Green Version]
- Balasubramanian, S.; Hurley, L.H.; Neidle, S. Targeting G-quadruplexes in gene promoters: A novel anticancer strategy? Nat. Rev. Drug Discov. 2011, 10, 261–275. [Google Scholar] [CrossRef]
- Cogoi, S.; Xodo, L.E. G-quadruplex formation within the promoter of the KRAS proto-oncogene and its effect on transcription. Nucleic Acids Res. 2006, 34, 2536–2549. [Google Scholar] [CrossRef]
- Catasti, P.; Chen, X.; Moyzis, R.K.; Bradbury, E.M.; Gupta, G. Structure-function correlations of the insulin-linked polymorphic region. J. Mol. Biol. 1996, 264, 534–545. [Google Scholar] [CrossRef] [PubMed]
- Brázda, V.; Hároníková, L.; Liao, J.C.C.; Fojta, M. DNA and RNA quadruplex-binding proteins. Int. J. Mol. Sci. 2014, 15, 17493–17517. [Google Scholar] [CrossRef]
- Gray, L.T.; Vallur, A.C.; Eddy, J.; Maizels, N. G quadruplexes are genomewide targets of transcriptional helicases XPB and XPD. Nat. Chem. Biol. 2014, 10, 313–318. [Google Scholar] [CrossRef]
- Paeschke, K.; Simonsson, T.; Postberg, J.; Rhodes, D.; Lipps, H.J. Telomere end-binding proteins control the formation of G-quadruplex DNA structures in vivo. Nat. Struct. Mol. Biol. 2005, 12, 847–854. [Google Scholar] [CrossRef] [PubMed]
- Mergny, J.-L.; Li, J.; Lacroix, L.; Amrane, S.; Chaires, J.B. Thermal difference spectra: A specific signature for nucleic acid structures. Nucleic Acids Res. 2005, 33, e138. [Google Scholar] [CrossRef]
- Fleming, A.M.; Zhou, J.; Wallace, S.S.; Burrows, C.J. A Role for the Fifth G-Track in G-Quadruplex Forming Oncogene Promoter Sequences during Oxidative Stress: Do These "Spare Tires" Have an Evolved Function? ACS Cent. Sci. 2015, 1, 226–233. [Google Scholar] [CrossRef] [PubMed]
- Dai, J.; Carver, M.; Punchihewa, C.; Jones, R.A.; Yang, D. Structure of the Hybrid-2 type intramolecular human telomeric G-quadruplex in K+ solution: Insights into structure polymorphism of the human telomeric sequence. Nucleic Acids Res. 2007, 35, 4927–4940. [Google Scholar] [CrossRef]
- Guédin, A.; Gros, J.; Alberti, P.; Mergny, J.-L. How long is too long? Effects of loop size on G-quadruplex stability. Nucleic Acids Res. 2010, 38, 7858–7868. [Google Scholar] [CrossRef] [Green Version]
- Tippana, R.; Xiao, W.; Myong, S. G-quadruplex conformation and dynamics are determined by loop length and sequence. Nucleic Acids Res. 2014, 42, 8106–8114. [Google Scholar] [CrossRef] [Green Version]
- Hildebrandt, M.; Lacombe, M.L.; Mesnildrey, S.; Véron, M. A human NDP-kinase B specifically binds single-stranded poly-pyrimidine sequences. Nucleic Acids Res. 1995, 23, 3858–3864. [Google Scholar] [CrossRef] [Green Version]
- Postel, E.H.; Berberich, S.J.; Flint, S.J.; Ferrone, C.A. Human c-myc transcription factor PuF identified as nm23-H2 nucleoside diphosphate kinase, a candidate suppressor of tumor metastasis. Science 1993, 261, 478–480. [Google Scholar] [CrossRef]
- Boissan, M.; Lacombe, M.L. Learning about the functions of NME/NM23: Lessons from knockout mice to silencing strategies. Naunyn-Schmiedeberg’s Arch. Pharmacol. 2011, 384, 421–431. [Google Scholar] [CrossRef]
- Yafe, A.; Etzioni, S.; Weisman-Shomer, P.; Fry, M. Formation and properties of hairpin and tetraplex structures of guanine-rich regulatory sequences of muscle-specific genes. Nucleic Acids Res. 2005, 33, 2887–2900. [Google Scholar] [CrossRef] [Green Version]
- Rehm, C.; Holder, I.T.; Groß, A.; Wojciechowski, F.; Urban, M.; Sinn, M.; Drescher, M.; Hartig, J.S. A bacterial DNA quadruplex with exceptional K + selectivity and unique structural polymorphism. Chem. Sci. 2014, 5, 2809–2818. [Google Scholar] [CrossRef]
- Kuryavyi, V.; Cahoon, L.A.; Seifert, H.S.; Patel, D.J. RecA-binding pilE G4 sequence essential for pilin antigenic variation forms monomeric and 5’ end-stacked dimeric parallel G-quadruplexes. Structure 2012, 20, 2090–2102. [Google Scholar] [CrossRef] [PubMed]
- Waller, Z.A.; Pinchbeck, B.J.; Buguth, B.S.; Meadows, T.G.; Richardson, D.J.; Gates, A.J. Control of bacterial nitrate assimilation by stabilization of G-quadruplex DNA. Chem. Commun. (Camb) 2016, 52, 13511–13514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piekna-Przybylska, D.; Sullivan, M.A.; Sharma, G.; Bambara, R.A. U3 region in the HIV-1 genome adopts a G-quadruplex structure in its RNA and DNA sequence. Biochemistry 2014, 53, 2581–2593. [Google Scholar] [CrossRef] [PubMed]
- Shin, Y.J.; Kumarasamy, V.; Camacho, D.; Sun, D. Involvement of G-quadruplex structures in regulation of human RET gene expression by small molecules in human medullary thyroid carcinoma TT cells. Oncogene 2015, 34, 1292–1299. [Google Scholar] [CrossRef]
- Kumarasamy, V.M.; Shin, Y.-J.; White, J.; Sun, D. Selective repression of RET proto-oncogene in medullary thyroid carcinoma by a natural alkaloid berberine. BMC Cancer 2015, 15, 599. [Google Scholar] [CrossRef]
- Guo, K.; Pourpak, A.; Beetz-Rogers, K.; Gokhale, V.; Sun, D.; Hurley, L.H. Formation of pseudosymmetrical G-quadruplex and i-motif structures in the proximal promoter region of the RET oncogene. J. Am. Chem. Soc. 2007, 129, 10220–10228. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Liu, W.-J.; Guo, K.; Rusche, J.J.; Ebbinghaus, S.; Gokhale, V.; Hurley, L.H. The proximal promoter region of the human vascular endothelial growth factor gene has a G-quadruplex structure that can be targeted by G-quadruplex-interactive agents. Mol. Cancer Ther. 2008, 7, 880–889. [Google Scholar] [CrossRef]
- Dexheimer, T.S.; Sun, D.; Hurley, L.H. Deconvoluting the structural and drug-recognition complexity of the G-quadruplex-forming region upstream of the bcl-2 P1 promoter. J. Am. Chem. Soc. 2006, 128, 5404–5415. [Google Scholar] [CrossRef] [PubMed]
- Suloway, C.; Pulokas, J.; Fellmann, D.; Cheng, A.; Guerra, F.; Quispe, J.; Stagg, S.; Potter, C.S.; Carragher, B. Automated molecular microscopy: The new Leginon system. J. Struct. Biol. 2005, 151, 41–60. [Google Scholar] [CrossRef]
- Punjani, A.; Rubinstein, J.L.; Fleet, D.J.; Brubaker, M.A. cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 2017, 14, 290–296. [Google Scholar] [CrossRef]
Sample Availability: Samples of ZmNDPK expression plasmid is available upon request from the authors. |











| Oligonucleotide | Sequence | CD | Tm | Competition % |
|---|---|---|---|---|
| A5UAD | GGGTtTTGAAGGGAGGAGtAtttAGGG | parallel | <30 | 73 |
| A5UAE | GGGTtTTGAAGGGAtGAGGAtttAGGG | anti-h | 37 | 16 |
| A5UAF | GGGTtTTGAAGGGAttAGGAGttAGGG | anti-h | ~30 | 15 |
| A5UAG | GGGTtTTGAAGGGAttAtGAGGtAGGG | anti-h | 40 | 40 |
| A5UAH (canonical) | GGGTtTTGAAGGGAttAttAGGGAGGG | parallel | 42 | 77 |
| A5UBF | GGGTtTTGAAtGGAGtAGGAGttAGGG | anti-h | <30 | 23 |
| A5UBG | GGGTtTTGAAtGGAGtAtGAGGtAGGG | anti-h | <30 | 50 |
| A5UBH | GGGTtTTGAAtGGAGtAttAGGGAGGG | parallel | <30 | 85 |
| A5UCF | GGGTtTTGAAttGAGGAGGAGttAGGG | parallel | <30 | 62 |
| A5UCG | GGGTtTTGAAttGAGGAtGAGGtAGGG | mixed | <30 | 36 |
| A5UCH | GGGTtTTGAAttGAGGAttAGGGAGGG | parallel | ~30 | 76 |
| A5UDH | GGGTtTTGAAtttAGGAGtAGGGAGGG | parallel | ~30 | 77 |
| A5UEH | GGGTtTTGAAtttAtGAGGAGGGAGGG | parallel | 35 | 81 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kopylov, M.; Jackson, T.M.; Stroupe, M.E. Bulged and Canonical G-Quadruplex Conformations Determine NDPK Binding Specificity. Molecules 2019, 24, 1988. https://doi.org/10.3390/molecules24101988
Kopylov M, Jackson TM, Stroupe ME. Bulged and Canonical G-Quadruplex Conformations Determine NDPK Binding Specificity. Molecules. 2019; 24(10):1988. https://doi.org/10.3390/molecules24101988
Chicago/Turabian StyleKopylov, Mykhailo, Trevia M. Jackson, and M. Elizabeth Stroupe. 2019. "Bulged and Canonical G-Quadruplex Conformations Determine NDPK Binding Specificity" Molecules 24, no. 10: 1988. https://doi.org/10.3390/molecules24101988
APA StyleKopylov, M., Jackson, T. M., & Stroupe, M. E. (2019). Bulged and Canonical G-Quadruplex Conformations Determine NDPK Binding Specificity. Molecules, 24(10), 1988. https://doi.org/10.3390/molecules24101988