Effect of Staple Age on DNA Origami Nanostructure Assembly and Stability
Abstract
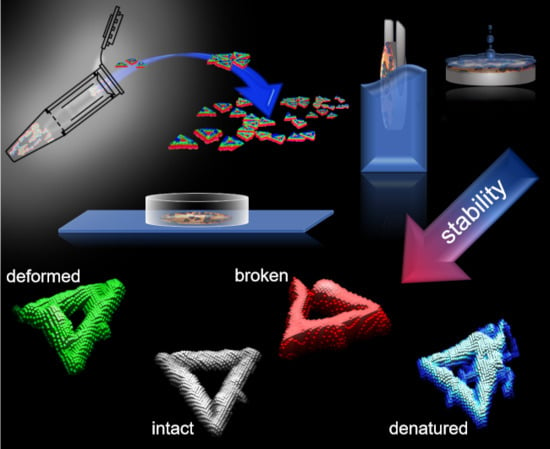
:1. Introduction
2. Results and Discussion
3. Materials and Methods
3.1. Preparation and Storage of the Staple Strands
3.2. DNA Origami Assembly and Purification
3.3. Preparation of DNA Origami Samples for AFM Analysis
3.4. AFM Imaging
3.5. Determination of Yields
3.6. MALDI-TOF Mass Spectrometry
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Nummelin, S.; Kommeri, J.; Kostiainen, M.A.; Linko, V. Evolution of Structural DNA Nanotechnology. Adv. Mater. 2018, 30, e1703721. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rothemund, P.W.K. Folding DNA to create nanoscale shapes and patterns. Nature 2006, 440, 297–302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hong, F.; Zhang, F.; Liu, Y.; Yan, H. DNA Origami: Scaffolds for Creating Higher Order Structures. Chem. Rev. 2017, 117, 12584–12640. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Jiang, Q.; Li, N.; Dai, L.; Liu, Q.; Song, L.; Wang, J.; Li, Y.; Tian, J.; Ding, B.; et al. DNA origami as an in vivo drug delivery vehicle for cancer therapy. ACS Nano 2014, 8, 6633–6643. [Google Scholar] [CrossRef] [PubMed]
- Douglas, S.M.; Bachelet, I.; Church, G.M. A logic-gated nanorobot for targeted transport of molecular payloads. Science 2012, 335, 831–834. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Jiang, Q.; Liu, S.; Zhang, Y.; Tian, Y.; Song, C.; Wang, J.; Zou, Y.; Anderson, G.J.; Han, J.-Y.; et al. A DNA nanorobot functions as a cancer therapeutic in response to a molecular trigger in vivo. Nat. Biotechnol. 2018, 36, 258–264. [Google Scholar] [CrossRef]
- Liu, J.; Song, L.; Liu, S.; Jiang, Q.; Liu, Q.; Li, N.; Wang, Z.-G.; Ding, B. A DNA-Based Nanocarrier for Efficient Gene Delivery and Combined Cancer Therapy. Nano Lett. 2018, 18, 3328–3334. [Google Scholar] [CrossRef]
- Smith, D.; Schüller, V.; Engst, C.; Rädler, J.; Liedl, T. Nucleic acid nanostructures for biomedical applications. Nanomedicine 2013, 8, 105–121. [Google Scholar] [CrossRef]
- Ochmann, S.E.; Vietz, C.; Trofymchuk, K.; Acuna, G.P.; Lalkens, B.; Tinnefeld, P. Optical Nanoantenna for Single Molecule-Based Detection of Zika Virus Nucleic Acids without Molecular Multiplication. Anal. Chem. 2017, 89, 13000–13007. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.; Nguyen, M.-K.; Natarajan, A.K.; Nguyen, V.H.; Kuzyk, A. A DNA Origami-Based Chiral Plasmonic Sensing Device. ACS Appl. Mater. Interfaces 2018, 10, 44221–44225. [Google Scholar] [CrossRef] [Green Version]
- Daems, D.; Pfeifer, W.; Rutten, I.; Saccà, B.; Spasic, D.; Lammertyn, J. Three-Dimensional DNA Origami as Programmable Anchoring Points for Bioreceptors in Fiber Optic Surface Plasmon Resonance Biosensing. ACS Appl. Mater. Interfaces 2018, 10, 23539–23547. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.; Kotthoff, L.; Olejko, L.; Resch-Genger, U.; Bald, I. DNA Origami-Based Förster Resonance Energy-Transfer Nanoarrays and Their Application as Ratiometric Sensors. ACS Appl. Mater. Interfaces 2018, 10, 23295–23302. [Google Scholar] [CrossRef] [PubMed]
- Aryal, B.R.; Westover, T.R.; Ranasinghe, D.R.; Calvopiña, D.G.; Uprety, B.; Harb, J.N.; Davis, R.C.; Woolley, A.T. Four-Point Probe Electrical Measurements on Templated Gold Nanowires Formed on Single DNA Origami Tiles. Langmuir 2018, 34, 15069–15077. [Google Scholar] [CrossRef] [PubMed]
- Bayrak, T.; Helmi, S.; Ye, J.; Kauert, D.; Kelling, J.; Schönherr, T.; Weichelt, R.; Erbe, A.; Seidel, R. DNA-Mold Templated Assembly of Conductive Gold Nanowires. Nano Lett. 2018, 18, 2116–2123. [Google Scholar] [CrossRef] [PubMed]
- Teschome, B.; Facsko, S.; Schönherr, T.; Kerbusch, J.; Keller, A.; Erbe, A. Temperature-Dependent Charge Transport through Individually Contacted DNA Origami-Based Au Nanowires. Langmuir 2016, 32, 10159–10165. [Google Scholar] [CrossRef] [PubMed]
- Uprety, B.; Jensen, J.; Aryal, B.R.; Davis, R.C.; Woolley, A.T.; Harb, J.N. Directional Growth of DNA-Functionalized Nanorods to Enable Continuous, Site-Specific Metallization of DNA Origami Templates. Langmuir 2017, 33, 10143–10152. [Google Scholar] [CrossRef] [PubMed]
- Kuzyk, A.; Schreiber, R.; Fan, Z.; Pardatscher, G.; Roller, E.-M.; Högele, A.; Simmel, F.C.; Govorov, A.O.; Liedl, T. DNA-based self-assembly of chiral plasmonic nanostructures with tailored optical response. Nature 2012, 483, 311–314. [Google Scholar] [CrossRef] [Green Version]
- Shen, X.; Song, C.; Wang, J.; Shi, D.; Wang, Z.; Liu, N.; Ding, B. Rolling up gold nanoparticle-dressed DNA origami into three-dimensional plasmonic chiral nanostructures. J. Am. Chem. Soc. 2012, 134, 146–149. [Google Scholar] [CrossRef]
- Prinz, J.; Schreiber, B.; Olejko, L.; Oertel, J.; Rackwitz, J.; Keller, A.; Bald, I. DNA Origami Substrates for Highly Sensitive Surface-Enhanced Raman Scattering. J. Phys. Chem. Lett. 2013, 4, 4140–4145. [Google Scholar] [CrossRef]
- Acuna, G.P.; Möller, F.M.; Holzmeister, P.; Beater, S.; Lalkens, B.; Tinnefeld, P. Fluorescence enhancement at docking sites of DNA-directed self-assembled nanoantennas. Science 2012, 338, 506–510. [Google Scholar] [CrossRef]
- Schreiber, R.; Luong, N.; Fan, Z.; Kuzyk, A.; Nickels, P.C.; Zhang, T.; Smith, D.M.; Yurke, B.; Kuang, W.; Govorov, A.O.; et al. Chiral plasmonic DNA nanostructures with switchable circular dichroism. Nat. Commun. 2013, 4, 2948. [Google Scholar] [CrossRef] [PubMed]
- Keller, A.; Bald, I.; Rotaru, A.; Cauët, E.; Gothelf, K.V.; Besenbacher, F. Probing electron-induced bond cleavage at the single-molecule level using DNA origami templates. ACS Nano 2012, 6, 4392–4399. [Google Scholar] [CrossRef] [PubMed]
- Voigt, N.V.; Tørring, T.; Rotaru, A.; Jacobsen, M.F.; Ravnsbaek, J.B.; Subramani, R.; Mamdouh, W.; Kjems, J.; Mokhir, A.; Besenbacher, F.; et al. Single-molecule chemical reactions on DNA origami. Nat. Nanotechnol. 2010, 5, 200–203. [Google Scholar] [CrossRef] [PubMed]
- Kielar, C.; Reddavide, F.V.; Tubbenhauer, S.; Cui, M.; Xu, X.; Grundmeier, G.; Zhang, Y.; Keller, A. Pharmacophore Nanoarrays on DNA Origami Substrates as a Single-Molecule Assay for Fragment-Based Drug Discovery. Angew. Chem. Int. Ed. Engl. 2018, 57, 14873–14877. [Google Scholar] [CrossRef] [PubMed]
- Endo, M.; Katsuda, Y.; Hidaka, K.; Sugiyama, H. A versatile DNA nanochip for direct analysis of DNA base-excision repair. Angew. Chem. Int. Ed. Engl. 2010, 49, 9412–9416. [Google Scholar] [CrossRef] [PubMed]
- Rackwitz, J.; Kopyra, J.; Dąbkowska, I.; Ebel, K.; Ranković, M.L.; Milosavljević, A.R.; Bald, I. Sensitizing DNA Towards Low-Energy Electrons with 2-Fluoroadenine. Angew. Chem. Int. Ed. Engl. 2016, 55, 10248–10252. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, S.; De, D.; Hidaka, K.; Kim, K.K.; Endo, M.; Sugiyama, H. Single molecule visualization and characterization of Sox2-Pax6 complex formation on a regulatory DNA element using a DNA origami frame. Nano Lett. 2014, 14, 2286–2292. [Google Scholar] [CrossRef] [PubMed]
- Ramakrishnan, S.; Subramaniam, S.; Stewart, A.F.; Grundmeier, G.; Keller, A. Regular Nanoscale Protein Patterns via Directed Adsorption through Self-Assembled DNA Origami Masks. ACS Appl. Mater. Interfaces 2016, 8, 31239–31247. [Google Scholar] [CrossRef]
- Shen, B.; Linko, V.; Tapio, K.; Kostiainen, M.A.; Toppari, J.J. Custom-shaped metal nanostructures based on DNA origami silhouettes. Nanoscale 2015, 7, 11267–11272. [Google Scholar] [CrossRef] [Green Version]
- Surwade, S.P.; Zhou, F.; Li, Z.; Powell, A.; O’Donnell, C.; Liu, H. Nanoscale patterning of self-assembled monolayers using DNA nanostructure templates. Chem. Commun. 2016, 52, 1677–1680. [Google Scholar] [CrossRef]
- Shen, B.; Linko, V.; Tapio, K.; Pikker, S.; Lemma, T.; Gopinath, A.; Gothelf, K.V.; Kostiainen, M.A.; Toppari, J.J. Plasmonic nanostructures through DNA-assisted lithography. Sci. Adv. 2018, 4, eaap8978. [Google Scholar] [CrossRef] [Green Version]
- Sajfutdinow, M.; Uhlig, K.; Prager, A.; Schneider, C.; Abel, B.; Smith, D.M. Nanoscale patterning of self-assembled monolayer (SAM)-functionalised substrates with single molecule contact printing. Nanoscale 2017, 9, 15098–15106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramakrishnan, S.; Krainer, G.; Grundmeier, G.; Schlierf, M.; Keller, A. Structural stability of DNA origami nanostructures in the presence of chaotropic agents. Nanoscale 2016, 8, 10398–10405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kielar, C.; Xin, Y.; Shen, B.; Kostiainen, M.A.; Grundmeier, G.; Linko, V.; Keller, A. On the Stability of DNA Origami Nanostructures in Low-Magnesium Buffers. Angew. Chem. Int. Ed Engl. 2018, 57, 9470–9474. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Surwade, S.P.; Powell, A.; O’Donnell, C.; Liu, H. Stability of DNA Origami Nanostructure under Diverse Chemical Environments. Chem. Mater. 2014, 26, 5265–5273. [Google Scholar] [CrossRef]
- Ramakrishnan, S.; Krainer, G.; Grundmeier, G.; Schlierf, M.; Keller, A. Cation-Induced Stabilization and Denaturation of DNA Origami Nanostructures in Urea and Guanidinium Chloride. Small 2017, 13. [Google Scholar] [CrossRef]
- Ramakrishnan, S.; Ijäs, H.; Linko, V.; Keller, A. Structural stability of DNA origami nanostructures under application-specific conditions. Comput. Struct. Biotechnol. J. 2018, 16, 342–349. [Google Scholar] [CrossRef]
- Hahn, J.; Wickham, S.F.J.; Shih, W.M.; Perrault, S.D. Addressing the instability of DNA nanostructures in tissue culture. ACS Nano 2014, 8, 8765–8775. [Google Scholar] [CrossRef]
- Wang, D.; Da, Z.; Zhang, B.; Isbell, M.A.; Dong, Y.; Zhou, X.; Liu, H.; Heng, J.Y.Y.; Yang, Z. Stability study of tubular DNA origami in the presence of protein crystallisation buffer. RSC Adv. 2015, 5, 58734–58737. [Google Scholar] [CrossRef]
- Bila, H.; Kurisinkal, E.E.; Bastings, M.M.C. Engineering a stable future for DNA-origami as a biomaterial. Biomater. Sci. 2019, 7, 532–541. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, P.; Xu, Y.; Li, X.; Zhu, Y.; Zhang, Y.; Zhu, J.; Huang, G.; He, D. Different Stability of DNA Origami Nanostructure between on Interface and in Bulk Solution. ACS Appl. Bio Mater. 2018, 1, 1424–1429. [Google Scholar] [CrossRef]
- Kroener, F.; Traxler, L.; Heerwig, A.; Rant, U.; Mertig, M. Magnesium-Dependent Electrical Actuation and Stability of DNA Origami Rods. ACS Appl. Mater. Interfaces 2019, 11, 2295–2301. [Google Scholar] [CrossRef]
- Engel, M.C.; Smith, D.M.; Jobst, M.A.; Sajfutdinow, M.; Liedl, T.; Romano, F.; Rovigatti, L.; Louis, A.A.; Doye, J.P.K. Force-Induced Unravelling of DNA Origami. ACS Nano 2018, 12, 6734–6747. [Google Scholar] [CrossRef]
- Fischer, S.; Hartl, C.; Frank, K.; Rädler, J.O.; Liedl, T.; Nickel, B. Shape and Interhelical Spacing of DNA Origami Nanostructures Studied by Small-Angle X-ray Scattering. Nano Lett. 2016, 16, 4282–4287. [Google Scholar] [CrossRef]
- Ramakrishnan, S.; Shen, B.; Kostiainen, M.A.; Grundmeier, G.; Keller, A.; Linko, V. Real-Time Observation of Superstructure-Dependent DNA Origami Digestion by DNase I Using High-Speed Atomic Force Microscopy. ChemBioChem 2019. [Google Scholar] [CrossRef]
- Zhu, B.; Zhao, Y.; Dai, J.; Wang, J.; Xing, S.; Guo, L.; Chen, N.; Qu, X.; Li, L.; Shen, J.; et al. Preservation of DNA Nanostructure Carriers: Effects of Freeze-Thawing and Ionic Strength during Lyophilization and Storage. ACS Appl. Mater. Interfaces 2017, 9, 18434–18439. [Google Scholar] [CrossRef]
- Linko, V.; Shen, B.; Tapio, K.; Toppari, J.J.; Kostiainen, M.A.; Tuukkanen, S. One-step large-scale deposition of salt-free DNA origami nanostructures. Sci. Rep. 2015, 5, 15634. [Google Scholar] [CrossRef] [Green Version]
- Wu, N.; Zhou, X.; Czajkowsky, D.M.; Ye, M.; Zeng, D.; Fu, Y.; Fan, C.; Hu, J.; Li, B. In situ monitoring of single molecule binding reactions with time-lapse atomic force microscopy on functionalized DNA origami. Nanoscale 2011, 3, 2481–2484. [Google Scholar] [CrossRef]
- Rajendran, A.; Endo, M.; Hidaka, K.; Tran, P.L.T.; Mergny, J.-L.; Sugiyama, H. Controlling the stoichiometry and strand polarity of a tetramolecular G-quadruplex structure by using a DNA origami frame. Nucleic Acids Res. 2013, 41, 8738–8747. [Google Scholar] [CrossRef] [Green Version]
- Chao, J.; Zhang, P.; Wang, Q.; Wu, N.; Zhang, F.; Hu, J.; Fan, C.H.; Li, B. Single-molecule imaging of DNA polymerase I (Klenow fragment) activity by atomic force microscopy. Nanoscale 2016, 8, 5842–5846. [Google Scholar] [CrossRef]
- Helmig, S.; Rotaru, A.; Arian, D.; Kovbasyuk, L.; Arnbjerg, J.; Ogilby, P.R.; Kjems, J.; Mokhir, A.; Besenbacher, F.; Gothelf, K.V. Single molecule atomic force microscopy studies of photosensitized singlet oxygen behavior on a DNA origami template. ACS Nano 2010, 4, 7475–7480. [Google Scholar] [CrossRef]
- Teshome, B.; Facsko, S.; Keller, A. Topography-controlled alignment of DNA origami nanotubes on nanopatterned surfaces. Nanoscale 2014, 6, 1790–1796. [Google Scholar] [CrossRef]
- Teschome, B.; Facsko, S.; Gothelf, K.V.; Keller, A. Alignment of Gold Nanoparticle-Decorated DNA Origami Nanotubes: Substrate Prepatterning versus Molecular Combing. Langmuir 2015, 31, 12823–12829. [Google Scholar] [CrossRef]
- Kielar, C.; Ramakrishnan, S.; Fricke, S.; Grundmeier, G.; Keller, A. Dynamics of DNA Origami Lattice Formation at Solid-Liquid Interfaces. ACS Appl. Mater. Interfaces 2018, 10, 44844–44853. [Google Scholar] [CrossRef]
- Gates, K.S. An overview of chemical processes that damage cellular DNA: Spontaneous hydrolysis, alkylation, and reactions with radicals. Chem. Res. Toxicol. 2009, 22, 1747–1760. [Google Scholar] [CrossRef]
- Jiranusornkul, S.; Laughton, C.A. Destabilization of DNA duplexes by oxidative damage at guanine: Implications for lesion recognition and repair. J. R. Soc. Interface 2008, 5 (Suppl. 3), 191–198. [Google Scholar] [CrossRef]
- Singh, S.K.; Szulik, M.W.; Ganguly, M.; Khutsishvili, I.; Stone, M.P.; Marky, L.A.; Gold, B. Characterization of DNA with an 8-oxoguanine modification. Nucleic Acids Res. 2011, 39, 6789–6801. [Google Scholar] [CrossRef] [Green Version]
- Röder, B.; Frühwirth, K.; Vogl, C.; Wagner, M.; Rossmanith, P. Impact of long-term storage on stability of standard DNA for nucleic acid-based methods. J. Clin. Microbiol. 2010, 48, 4260–4262. [Google Scholar] [CrossRef]
Sample Availability: Not available. |





| AFM Image | N(DNA origami) | Intact DNA Origami (%) | Broken DNA Origami (%) | Denatured DNA Origami (%) | Deformed DNA Origami (%) |
|---|---|---|---|---|---|
| 1 | 524 | 89.5 | 4.2 | 4.8 | 1.5 |
| 2 | 518 | 88.2 | 6.2 | 4.1 | 1.5 |
| 3 | 444 | 86.0 | 5.0 | 5.2 | 3.8 |
| 4 | 515 | 86.8 | 4.8 | 6.0 | 3.1 |
| 5 | 491 | 88.0 | 5.7 | 3.1 | 3.3 |
| 6 | 526 | 85.2 | 7.0 | 3.4 | 4.4 |
| 7 | 552 | 88.2 | 4.5 | 3.1 | 4.2 |
| 8 | 554 | 87.5 | 6.0 | 2.7 | 3.8 |
| Sum | 4124 | 87.4 ± 1.4 | 5.4 ± 1.0 | 4.0 ± 1.2 | 3.2 ± 1.1 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kielar, C.; Xin, Y.; Xu, X.; Zhu, S.; Gorin, N.; Grundmeier, G.; Möser, C.; Smith, D.M.; Keller, A. Effect of Staple Age on DNA Origami Nanostructure Assembly and Stability. Molecules 2019, 24, 2577. https://doi.org/10.3390/molecules24142577
Kielar C, Xin Y, Xu X, Zhu S, Gorin N, Grundmeier G, Möser C, Smith DM, Keller A. Effect of Staple Age on DNA Origami Nanostructure Assembly and Stability. Molecules. 2019; 24(14):2577. https://doi.org/10.3390/molecules24142577
Chicago/Turabian StyleKielar, Charlotte, Yang Xin, Xiaodan Xu, Siqi Zhu, Nelli Gorin, Guido Grundmeier, Christin Möser, David M. Smith, and Adrian Keller. 2019. "Effect of Staple Age on DNA Origami Nanostructure Assembly and Stability" Molecules 24, no. 14: 2577. https://doi.org/10.3390/molecules24142577
APA StyleKielar, C., Xin, Y., Xu, X., Zhu, S., Gorin, N., Grundmeier, G., Möser, C., Smith, D. M., & Keller, A. (2019). Effect of Staple Age on DNA Origami Nanostructure Assembly and Stability. Molecules, 24(14), 2577. https://doi.org/10.3390/molecules24142577