Rapid Screening of Forskolin-Type Diterpenoids of Blumea aromatica DC Using Ultra-High-Performance Liquid Chromatography Tandem Quadrupole Time-Of-Flight Mass Spectrometry Based on the Mass Defect Filtering Approach
Abstract
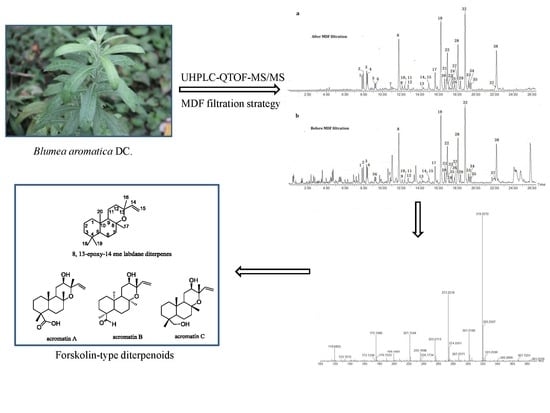
:1. Introduction
2. Results
2.1. Optimization of Sample Preparation and UHPLC-QTOF-MS Analysis Conditions
2.2. Establishment of the MDF Filtration Method
2.3. FSKD Analogue Identification After Filtration Using the MDF Approach
2.3.1. Structural Elucidation of Acromatin A, Acromatin B, and Acromatin C
2.3.2. Structural Elucidation of Forskolin
2.4. Accuracy Evaluation of the MDF Filtration Method
3. Materials and Methods
3.1. Chemicals and Materials
3.2. Preparation of Sample and Standard Solutions
3.3. UHPLC-QTOF/MS Conditions
3.4. Data Process
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| UHPLC | Ultra-high |
| QToF | Quadrupole Time of Flight |
| MDF | Mass defect filter |
| FSKD | forskolin-type diterpenoids |
| TCM | Traditional Chinese medicines |
References
- Zhang, L.; Shen, H.; Xu, J.; Xu, J.D.; Li, Z.L.; Wu, J.; Zou, J.T.; Wu, J.; Liu, L.F.; Li, S.L. UPLC-QTOF-MS/MS-guided isolation and purification of sulfur-containing derivatives from sulfur-fumigated edible herbs, a case study on ginseng. Food Chem. 2018, 246, 202–210. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.L.; Lin, H.Q.; Tan, J.; Wang, C.Z.; Wang, H.; Wu, F.L.; Dong, Q.H.; Liu, Y.H.; Li, P.Y.; Liu, J.P. UPLC-QTOF/MS-Based Nontargeted Metabolomic Analysis of Mountain- and Garden-Cultivated Ginseng of Different Ages in Northeast China. Molecules 2019, 24, 33. [Google Scholar] [CrossRef] [PubMed]
- He, L.L.; Zhang, Z.F.; Liu, Y.; Chen, D.Q.; Yuan, M.H.; Dong, G.T.; Luo, P.; Yan, Z.G. Rapid discrimination of raw and sulfur-fumigated Smilax glabra based on chemical profiles by UHPLC-QTOF-MS/MS coupled with multivariate statistical analysis. Food Res. Int. 2018, 108, 226–236. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.H.; Meng, Y.H.; Zhai, C.M.; Wang, M.; Avula, B.; Yuk, J.; Smith, K.M.; Isaac, G.; Khan, I.A. The Chemical Characterization of Eleutherococcus senticosus and Ci-wu-jia Tea Using UHPLC-UV-QTOF/MS. Int. J. Mol. Sci. 2019, 20, 475. [Google Scholar] [CrossRef]
- Rivera, G.A.; Vivas, D.B.; Alfonso, F.P.; Ibañez, E.; Cifuentes, A. Recent applications of high resolution mass spectrometry for the characterization of plant natural products. Trends Anal. Chem. 2019, 112, 87–101. [Google Scholar] [CrossRef]
- Pan, H.Q.; Yao, C.L.; Yang, W.Z.; Yao, S.; Huang, Y.; Zhang, Y.B.; Wu, W.Y.; Guo, D.A. An enhanced strategy integrating offline two-dimensional separation and step-wise precursor ion list-based raster-mass defect filter: Characterization of indole alkaloids in five botanical origins of Uncariae Ramulus Cum Unicis as an exemplary application. J. Chromatogr. A 2018, 1563, 124–134. [Google Scholar] [CrossRef]
- Zhou, W.; Shan, J.; Meng, M. A two-step ultra-high-performance liquid chromatography-quadrupole/time of flight mass spectrometry with mass defect filtering method for rapid identification of analogues from known components of different chemical structure types in Fructus Gardeniae-Fructus Forsythiae herb pair extract and in rat’s blood. J. Chromatogr. A 2018, 1563, 99–123. [Google Scholar] [CrossRef]
- Yang, B.; Li, H.; Ruan, Q.F.; Xue, Y.Y.; Cao, D.; Zhou, X.H.; Jiang, S.Q.; Yi, T.; Jin, J.; Zhao, Z.X. A facile and selective approach to the qualitative and quantitative analysis of triterpenoids and phenylpropanoids by UPLC/Q-TOF-MS/MS for the quality control of Ilex rotunda. J. Pharm. Biomed. Anal. 2018, 157, 44–58. [Google Scholar] [CrossRef]
- Zhao, W.J.; Shang, Z.P.; Li, Q.Q.; Huang, M.R.; He, W.B.; Wang, Z.B.; Zhang, J.Y. Rapid Screening and Identification of Daidzein Metabolites in Rats Based on UHPLC-LTQ-Orbitrap Mass Spectrometry Coupled with Data-Mining Technologies. Molecules 2018, 23, 151. [Google Scholar] [CrossRef]
- Ekanayaka, E.A.; Celiz, M.D.; Jones, A.D. Relative mass defect filtering of mass spectra: A path to discovery of plant specialized metabolites. Plant Physiol. 2015, 167, 1221–1232. [Google Scholar] [CrossRef]
- Asada, Y.; Li, W.; Terada, T.; Kuang, X.; Li, Q.; Yoshikawa, T.; Hamaguchi, S.; Namekata, I.; Tanaka, H.; Koike, K. Labdane-type diterpenoids from hairy root cultures of Coleus forskohlii, possible intermediates in the biosynthesis of forskolin. Phytochemistry 2012, 79, 141–146. [Google Scholar] [CrossRef] [PubMed]
- Alasbahi, R.H.; Melzig, M.F. Forskolin and derivatives as tools for studying the role of cAMP. Pharmazie 2012, 67, 5–13. [Google Scholar] [CrossRef] [PubMed]
- Ríos-Silva, M.; Trujillo, X.; Trujillo-Hernández, B.; Sánchez-Pastor, E.; Urzúa, Z.; Mancilla, E.; Huerta, M. Effect of chronic administration of forskolin on glycemia and oxidative stress in rats with and without experimental diabetes. Int. J. Med. Sci. 2014, 11, 448–452. [Google Scholar] [CrossRef] [PubMed]
- Sapio, L.; Gallo, M.; Illiano, M.; Chiosi, E.; Naviglio, D.; Spina, A.; Naviglio, S. The Natural cAMP Elevating Compound Forskolin in Cancer Therapy: Is It Time? J. Cell Physiol. 2017, 232, 922–927. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.L.; Lou, C.Y.; Ma, N. Forskolin exerts anticancer roles in non-Hodgkin’s lymphomas via regulating Axin/β-catenin signaling pathway. Cancer Manag. Res. 2019, 11, 1685–1696. [Google Scholar] [CrossRef] [PubMed]
- Du, X.; Shi, R.; Wang, Y.; Wu, W.; Sun, S.; Dai, Z.; Chen, C.; Weng, Z.; Li, X.; Liu, Q.; et al. Isoforskolin and forskolin attenuate lipopolysaccharide-induced inflammation through TLR4/MyD88/NF-κB cascades in human mononuclear leukocytes. Phytother. Res. 2019, 33, 602–609. [Google Scholar] [CrossRef]
- Chen, Y.; Wen, J.G.; Feng, J.J.; Wang, Y.H.; Li, T.F.; Nurmi, K.; Eklund, K.K.; Xing, D. Forskolin attenuates the NLRP3 inflammasome activation and IL-1β secretion in human macrophages. Pediatr. Res. 2019, 1. [Google Scholar] [CrossRef]
- Bodiwala, H.S.; Sabde, S.; Mitra, D.; Bhutani, K.K.; Singh, I.P. Anti-HIV diterpenes from Coleus forskohlii. Nat. Prod. Commun. 2009, 4, 1173–1175. [Google Scholar] [CrossRef]
- Kavitha, C.; Rajamani, K.; Vadivel, E. Coleus forskohlii: A comprehensive review on morphology, phytochemistry and pharmacological aspects. J. Med. Plants Res. 2010, 4, 278–285. [Google Scholar]
- Lan, M.S.; Luo, C.; Tan, C.H.; Chen, L.; Wei, S.; Zhu, D.Y. Study on chemical constituents of the ethyl acetate extract from Blumea aromatica. J. Chin. Med. Mater. 2012, 35, 229–231. [Google Scholar]
- Xu, L.; Lu, J.; Li, W.; Kong, L. Studies on the chemical constituents in root of Coleus forskohlii. China J. Chin. Mater. Med. 2005, 30, 1753–1755. [Google Scholar]
- Shan, Y.; Xu, L.; Lu, Y.; Wang, X.; Zheng, Q.; Kong, L.; Niwa, M. Diterpenes from Coleus forskohlii (WILLD.) BRIQ. (Labiatae). Chem. Pharm. Bull. 2008, 56, 52–56. [Google Scholar] [CrossRef]
- Tan, T.; Luo, Y.; Zhong, C.C.; Xu, X.; Feng, Y.L. Comprehensive profiling and characterization of coumarins from roots stems, leaves, branches, and seeds of Chimonanthus nitens Oliv. using ultra-performance liquid chromatography/quadrupole-time-of-flight mass spectrometry combined with modified mass defect filter. J. Pharm. Biomed. Anal. 2017, 141, 140–148. [Google Scholar] [CrossRef]
- Fu, L.L.; Ding, H.; Han, L.F.; Jia, L.; Yang, W.Z.; Zhang, C.X.; Hu, Y.; Zuo, T.T.; Gao, X.M.; Guo, D.A. Simultaneously targeted and untargeted multicomponent characterization of Erzhi Pill by offline two-dimensional liquid chromatography/quadrupole-Orbitrap mass spectrometry. J. Chromatogr. A 2019, 1584, 87–96. [Google Scholar] [CrossRef]
- Ren, D.B.; Ran, L.; Yang, C.; Xu, M.L.; Yi, L.Z. Integrated strategy for identifying minor components in complex samples combining mass defect, diagnostic ions and neutral loss information based on ultra-performance liquid chromatography-high resolution mass spectrometry platform: Folium Artemisiae Argyi as a case study. J. Chromatogr. A 2018, 1550, 34–35. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds acromatin A, acromatin B, and acromatin C are available from the authors. |







| Peak No. | Retention Time (min) | ESI−, m/z * | ESI+, m/z | Formula | MS/MS Fragment Ions in Positive Mode ** | |
|---|---|---|---|---|---|---|
| [M − H]− | [M + COOH]− | |||||
| 1 | 7.81 | 369.2282 | 415.2367 | 388.2722 [M + H2O]+ | C20H34O6 | 353.2346, 335.2242, 317.2135, 305.2133, 299.2022, 289.2181, 287.2037, 275.2031, 271.2077, 259.2080, 253.1980, 237.1858, 229.1974, 221.1555 |
| 393.2276 [M + Na]+ | ||||||
| 741.4863 [2M + H]+ | ||||||
| 2 | 7.95 | 369.2274 | 415.2335 | 393.2271 [M + Na]+ | C20H34O6 | 353.2341, 335.2237, 317.2133, 299.2029, 289.2183, 287.2025, 275.2028, 271.2073, 259.2076, 255.2122, 253.1950, 221.1556 |
| 741.4832 [2M + H]+ | ||||||
| 763.4637 [2M + Na]+ | ||||||
| 3 | 8.30 | 369.2273 | 415.2336 | 388.2710 [M + H2O]+ | C20H34O6 | 353.2343, 335.2233, 317.2122, 305.2129, 299.2018, 289.2175, 287.2021, 275.2022, 271.2075, 259.2068, 253.1958, 229.1965, 221.1552 |
| 393.2259 [M + Na]+ | ||||||
| 741.4811 [2M + H]+ | ||||||
| 4 | 8.40 | 369.2279 | 415.2338 | 388.2710 [M + H2O]+ | C20H34O6 | 353.2345, 335.2234, 317.2120, 305.2132, 299.2014, 289.2176, 287.2024, 275.2027, 271.2075, 259.2065, 253.1952, 229.1965, 221.1556 |
| 393.2260 [M + Na]+ | ||||||
| 741.4801 [2M + H]+ | ||||||
| 763.4626 [2M + Na]+ | ||||||
| 5 | 9.19 | 351.2180 | 397.2241 | 370.2602 [M + H2O]+ | C20H32O5 | 335.2232, 317.2122, 305.2121, 299.2022, 289.2179, 287.2014, 275.2006, 271.2074, 255.2113, 253.1974, 221.1557, 175.1492 |
| 375.2152 [M + Na]+ | ||||||
| 705.4606 [2M + H]+ | ||||||
| 727.4465 [2M + Na]+ | ||||||
| 6 | 9.29 | 351.2178 | 397.2248 | 370.2602 [M + H2O]+ | C20H32O5 | 335.2232, 317.2122, 305.2121, 303.2323, 299.2022, 289.2179, 287.2021, 275.2006, 271.2074, 259.2070, 253.1974, 221.1557, 175.1491 |
| 375.2159 [M + Na]+ | ||||||
| 705.4611 [2M + H]+ | ||||||
| 727.4465 [2M + Na]+ | ||||||
| 7 | 10.93 | 351.2176 | 397.2245 | 370.2604 [M + H2O]+ | C20H32O5 | 335.2235, 317.2125, 305.2120, 303.2328, 299.2020, 289.2176, 275.2010, 271.2071, 259.2073, 253.1970, 221.1553 |
| 375.2155 [M + Na]+ | ||||||
| 705.4610 [2M + H]+ | ||||||
| 727.4466 [2M + Na]+ | ||||||
| 8 | 11.74 | 307.1905 | 353.1953 | 309.2070 [M + H]+ | C18H28O4 | 291.1969, 273.1863, 263.2017, 255.1757, 245.1913, 227.1808, 221.1551 |
| 326.2342 [M + H2O]+ | ||||||
| 331.1891 [M + Na]+ | ||||||
| 9 | 12.12 | 353.233 | 399.2389 | 377.2291 [M + Na]+ | C20H34O5 | 337.2320, 319.2272, 301.2164, 289.2163, 273.2215, 271.2060, 253.1949, 221.1548, 175.1482 |
| 10 | 12.36 | 353.233 | 399.2389 | --- | C20H34O5 | 337.2322, 319.2274, 301.2160, 289.2165, 273.2210, 271.2061, 253.1946, 221.1545, 175.1484 |
| 11 | 12.38 | 365.1948 | 411.2001 | --- | C20H30O6 | 349.2005, 319.2261, 301.2149, 273.2208, 255.2085, 221.1521, 175.1489 |
| 12 | 12.76 | 353.2316 | 399.2379 | --- | C20H34O5 | 337.2365, 319.2250, 301.2152, 289.2156, 279.1592, 273.2215, 259.2047, 255.2110, 253.1702, 221.1533, 175.1490 |
| 13 | 14.36 | 353.2319 | 399.2379 | 377.2271 [M + Na]+ | C20H34O5 | 337.2360, 319.2256, 301.2150, 289.2159, 279.1590, 273.2212, 259.2053, 255.2111, 253.1703, 221.1535, 175.1488 |
| 14 | 14.96 | 335.2217 | 381.2268 | --- | C20H32O4 | 319.2276, 301.2173, 287.2014, 289.2154, 273.2222, 269.1906, 255.2115, 221.1545 |
| 15 | 14.98 | 349.2011 | 395.2065 | --- | C20H30O5 | 333.2071, 315.1973, 287.2030, 269.1908, 255.2138, 227.1819, 221.1565 |
| 16 | 15.28 | --- | --- | 634.5107 [2M + H2O]+ | C19H32O3 | 291.2344, 273.2238, 255.2130, 207.1768, 199.1505, 191.1453, 175.1503 |
| 17 | 15.63 | 335.2231 | 381.2283 | --- | C20H32O4 | 319.2298, 301.2192, 283.2084, 273.2283, 255.2133, 221.1560, 207.1770, 189.1659, 175.1502 |
| 18 | 16.25 | 333.2078 | 379.2132 | 335.2216 [M + H]+ | C20H30O4 | 317.2112, 299.2007, 289.2163, 271.2057, 253.1952, 243.2113, 221.1542, 175.1483, 161.1327, 145.1014 |
| 357.2035 [M + Na]+ | ||||||
| 331.2238 [M + Na]+ | ||||||
| 639.4576 [2M + Na]+ | ||||||
| 19 | 16.43 | --- | --- | 331.2238 [M + Na]+ | C19H32O3 | 291.2314, 273.2210, 255.2104, 207.1739, 199.1477, 191.1429, 175.1480 |
| 639.4576 [2M + Na]+ | ||||||
| 20 | 16.65 | 335.2207 | 381.2254 | 359.2176 [M + Na]+ | C20H32O4 | 319.2257, 301.2152, 283.2047, 273.2204, 255.2104, 221.1533 |
| 21# | 16.87 | 321.2061 | 367.2113 | --- | C19H30O4 | 305.2112, 287.2007, 259.2059, 241.1946, 231.2101, 221.1540, 175.1487 |
| 22 | 16.94 | 333.2064 | 379.2114 | 335.2225 [M + H]+ | C20H30O4 | 317.2118, 303.2327, 299.2014, 285.2218, 271.2065, 253.1960, 221.1547, 207.1751, 175.1487 |
| 357.2048 [M + Na]+ | ||||||
| 23 | 17.03 | 335.2215 | 381.2254 | 337.2387 [M + H]+ | C20H32O4 | 319.2274, 301.2170, 289.2174, 273.2222, 255.2122, 221.1542, 193.1231, 175.1484 |
| 673.4677 [2M + H]+ | ||||||
| 24# | 17.16 | 349.2016 | 395.2074 | --- | C20H30O5 | 333.2086, 315.1972, 287.2021, 269.1918, 247.1706, 243.2122, 235.1702, 221.1907, 203.1807 |
| 25 | 17.46 | 335.2232 | 381.2286 | 337.2384 [M + H]+ | C20H32O4 | 319.2277, 301.2173, 289.2173, 273.2225, 255.2121, 221.1546, 207.1761, 199.1497, 193.1228, 175.1493 |
| 354.2652 [M + H2O]+ | ||||||
| 26 | 17.73 | 347.1849 | 393.1899 | 349.2000 [M + H]+ | C20H28O5 | 303.1945, 285.1851, 274.2733, 240.2318, 221.1531, 207.1741, 175.1471 |
| 27 | 18.01 | 335.2225 | 381.2278 | 359.2196 [M + Na]+ | C20H32O4 | 319.2268, 301.6121, 273.2212, 255.2112, 221.1540, 199.1494, 175.1490 |
| 695.4498 [2M + Na]+ | ||||||
| 28# | 18.08 (acromatin C) | 335.2226 | 381.2281 | 690.4946 [2M + H2O]+ | C20H32O4 | 319.2274, 301.2170, 273.2220, 255.2115, 235.1695, 221.1543, 199.1485, 193.1228, 175.1493 |
| 29 | 18.35 | 335.2228 | 381.228 | 359.2162 [M + Na]+ | C20H32O4 | 319.2274, 301.2170, 273.2220, 255.2115, 235.1698, 221.1543, 207.1731, 175.1493 |
| 30 | 18.65 | --- | --- | 345.2425 [M + Na]+ 667.4901 [2M + Na]+ | C20H34O3 | 305.2476, 287.2370, 275.2368, 269.2266, 257.2272, 207.1748, 189.1643, 177.1639 |
| 31 | 18.79 | --- | --- | 662.5363 [2M + H2O]+ | C20H34O3 | 305.2482, 287.2376, 275.2368, 269.2271,257.2288, 229.1962, 207.1753, 189.1646, 177.1646, 161.1337, 149.1328 |
| 32 | 18.84 | 335.2231 | 381.2287 | 359.2205 [M + Na]+ 690.4948 [2M + H2O]+ 695.4514 [2M + Na]+ | C20H32O4 | 319.2279, 305.2486, 301.2169, 287.2375, 273.2223, 255.2117, 221.1548, 207.1755, 199.1490, 189.1647, 175.1492 |
| 33 | 19.20 | 349.2385 | 395.2438 | 368.2815 [M + H2O]+ 718.5308 [2M + H2O]+ | C21H34O4 | 333.2448, 319.2286, 301.2179, 283.2076, 273.2235, 255.2127, 235.1706, 221.1554, 207.1403, 193.1242, 175.1493, 161.1339 |
| 34 | 19.31 | 349.2379 | 395.2432 | --- | C21H34O4 | 335.2238, 319.2288, 301.2180, 289.2183, 283.2069, 273.2222, 255.2126, 221.1554, 207.1403, 193.1242, 175.1493, 161.1339 |
| 35 | 19.45 | 335.2223 | 381.2276 | 359.2200 [M + Na]+ 673.4678 [2M + H]+ | C20H32O4 | 319.2267, 301.2168, 291.2322, 273.2218, 255.2110, 221.1540, 207.1749, 203.1799, 189.1640, 175.1494 |
| 36# | 19.52 (acromatin B) | --- | --- | 345.2401 [M + Na]+ 667.4901 [2M + Na]+ | C20H34O3 | 305.2476, 287.2370, 275.2368, 269.2266, 257.2271, 207.1748, 189.1643, 177.1639, 161.1328, 149.1325 |
| 37# | 20.17 (acromatin A) | --- | --- | 658.5023 [2M + H2O]+ 663.4627 [2M + Na]+ | C20H32O3 | 303.2314, 285.2208, 275.2384, 257.2264, 205.1589, 201.1648, 187.1484, 177.1638, 149.1330 |
| 38 | 22.20 | 319.2278 | 365.2329 | --- | C20H32O3 | 303.2325, 285.2217, 257.2268, 233.1548, 221.1542, 201.1646, 175.1483, 163.1487, 147.1175 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, L.; Zhang, Z.; Yao, C.; Miao, J.; Yan, B.; Wu, L.; Pan, L.; Song, Z.; Wei, S. Rapid Screening of Forskolin-Type Diterpenoids of Blumea aromatica DC Using Ultra-High-Performance Liquid Chromatography Tandem Quadrupole Time-Of-Flight Mass Spectrometry Based on the Mass Defect Filtering Approach. Molecules 2019, 24, 3073. https://doi.org/10.3390/molecules24173073
He L, Zhang Z, Yao C, Miao J, Yan B, Wu L, Pan L, Song Z, Wei S. Rapid Screening of Forskolin-Type Diterpenoids of Blumea aromatica DC Using Ultra-High-Performance Liquid Chromatography Tandem Quadrupole Time-Of-Flight Mass Spectrometry Based on the Mass Defect Filtering Approach. Molecules. 2019; 24(17):3073. https://doi.org/10.3390/molecules24173073
Chicago/Turabian StyleHe, Lili, Zhifeng Zhang, Caiyun Yao, Jianhua Miao, Bingxiong Yan, Lingling Wu, Limei Pan, Zhijun Song, and Shugen Wei. 2019. "Rapid Screening of Forskolin-Type Diterpenoids of Blumea aromatica DC Using Ultra-High-Performance Liquid Chromatography Tandem Quadrupole Time-Of-Flight Mass Spectrometry Based on the Mass Defect Filtering Approach" Molecules 24, no. 17: 3073. https://doi.org/10.3390/molecules24173073
APA StyleHe, L., Zhang, Z., Yao, C., Miao, J., Yan, B., Wu, L., Pan, L., Song, Z., & Wei, S. (2019). Rapid Screening of Forskolin-Type Diterpenoids of Blumea aromatica DC Using Ultra-High-Performance Liquid Chromatography Tandem Quadrupole Time-Of-Flight Mass Spectrometry Based on the Mass Defect Filtering Approach. Molecules, 24(17), 3073. https://doi.org/10.3390/molecules24173073