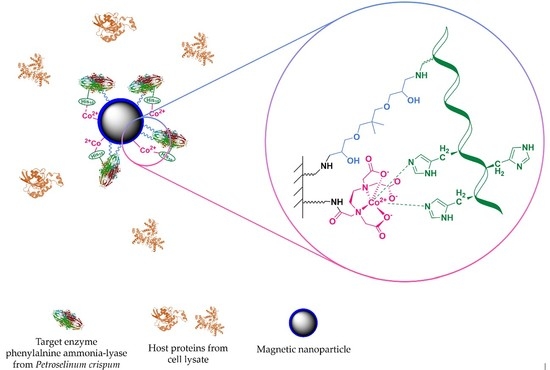
“Fishing and Hunting”—Selective Immobilization of a Recombinant Phenylalanine Ammonia-Lyase from Fermentation Media
Abstract
:1. Introduction
1.1. Importance of Biocatalysts
1.1.1. Whole Cells and Isolated Enzymes as Biocatalysts
1.1.2. Biocatalyst Production by Fermentation, Recombinant Techniques
1.1.3. Downstream Processes for Enzyme Production
1.2. Improved Immobilization Techniques for Biocatalysts: Methods, Pros and Cons
1.2.1. Whole-Cell Immobilization Strategies
1.2.2. Strategies for Immobilization of Cell-Free Forms of Enzymes
1.3. Immobilization Methods of Isolated Enzymes Simplifying the Downstream Process
1.3.1. Embedding as Purification Free Process
1.3.2. Affinity-Based Methods
2. Results and Discussion
2.1. Optimizing the Epoxy-Chelate Ratio
2.2. Comparison of the Biocatalytic Activity of Simple Epoxy – and Mixed Epoxide/EDa Functionalized MNPs
2.3. Effect of the Nature of Multiepoxide Agents on the MNPs for the Immobilization of PcPAL
2.4. Operational Stability of the Immobilized PcPAL Biocatalysts
3. Materials and Methods
3.1. Materials and Analysis
3.2. Expression and Purification of PcPAL
3.3. Surface Treatment of Aminopropylsilane-Coated Magnetic Nanoparticles with EDa and Multiepoxides at Different Ratios
3.4. Metal Ion Complexation of the Surface-Treated MNPs and Immobilization of PcPAL
3.5. Activity of the Immobilized PcPAL Biocatalysts in the Ammonia Elimination of l-Phenylalanine
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Woodley, J.M. Accelerating the implementation of biocatalysis in industry. Appl. Microbiol. Biotechnol. 2019, 103, 4733–4739. [Google Scholar] [CrossRef] [PubMed]
- Chapman, J.; Ismail, A.E.; Dinu, C.Z. Industrial Applications of Enzymes: Recent Advances, Techniques, and Outlooks. Catalysts 2018, 8, 238. [Google Scholar] [CrossRef]
- Devine, P.N.; Howard, R.M.; Kumar, R.; Thompson, M.P.; Truppo, M.D.; Turner, N.J. Extending the application of biocatalysis to meet the challenges of drug development. Nat. Rev. Chem. 2018, 2, 409–421. [Google Scholar] [CrossRef]
- Choi, J.-M.; Han, S.-S.; Kim, H.-S. Industrial applications of enzyme biocatalysis: Current status and future aspects. Biotechnol. Adv. 2015, 33, 1443–1454. [Google Scholar] [CrossRef] [PubMed]
- Ghanem, A. Trends in lipase-catalyzed asymmetric access to enantiomerically pure/enriched compounds. Tetrahedron 2007, 63, 1721–1754. [Google Scholar] [CrossRef]
- de Miranda, A.S.; Miranda, L.S.M.; de Souza, R.O.M.A. Lipases: Valuable catalysts for dynamic kinetic resolutions. Biotechnol. Adv. 2015, 33, 372–393. [Google Scholar] [CrossRef]
- Zhang, X.-Y.; Xu, Z.-H.; Zong, M.-H.; Wang, C.-F.; Li, N. Selective Synthesis of Furfuryl Alcohol from Biomass-Derived Furfural Using Immobilized Yeast Cells. Catalysts 2019, 9, 70. [Google Scholar] [CrossRef]
- Martín-Matute, B.; Bäckvall, J.-E. Dynamic kinetic resolution catalyzed by enzymes and metals. Curr. Opin. Chem. Biol. 2007, 11, 226–232. [Google Scholar] [CrossRef]
- Zhu, S.; Zheng, G. Dynamic kinetic resolution of Vince lactam catalyzed by γ-lactamases: A mini-review. J. Ind. Microbiol. Biotechnol. 2018, 45, 1017–1031. [Google Scholar] [CrossRef]
- Tian, W.; Sun, C.; Zheng, M.; Harmer, J.R.; Yu, M.; Zhang, Y.; Peng, H.; Zhu, D.; Deng, Z.; Chen, S.-L.; et al. Efficient biosynthesis of heterodimeric C3-aryl pyrroloindoline alkaloids. Nat. Commun. 2018, 9, 4428. [Google Scholar] [CrossRef]
- Venkataraman, H.; te Poele, E.M.; Rosłoniec, K.Z.; Vermeulen, N.; Commandeur, J.N.M.; van der Geize, R.; Dijkhuizen, L. Biosynthesis of a steroid metabolite by an engineered Rhodococcus erythropolis strain expressing a mutant cytochrome P450 BM3 enzyme. Appl. Microbiol. Biotechnol. 2015, 99, 4713–4721. [Google Scholar] [CrossRef] [PubMed]
- Putkaradze, N.; Kiss, F.M.; Schmitz, D.; Zapp, J.; Hutter, M.C.; Bernhardt, R. Biotransformation of prednisone and dexamethasone by cytochrome P450 based systems – Identification of new potential drug candidates. J. Biotechnol. 2017, 242, 101–110. [Google Scholar] [CrossRef] [PubMed]
- Xiong, S.; Wang, Y.; Yao, M.; Liu, H.; Zhou, X.; Xiao, W.; Yuan, Y. Cell foundry with high product specificity and catalytic activity for 21-deoxycortisol biotransformation. Microb. Cell Fact. 2017, 16, 105. [Google Scholar] [CrossRef] [PubMed]
- Morlock, L.K.; Grobe, S.; Balke, K.; Mauersberger, S.; Böttcher, D.; Bornscheuer, U.T. Protein Engineering of the Progesterone Hydroxylating P450-Monooxygenase CYP17A1 Alters Its Regioselectivity. ChemBioChem 2018, 19, 1954–1958. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Han, K.; Kim, M.-J.; Park, J. Chemoenzymatic Dynamic Kinetic Resolution of Alcohols and Amines. European J. Org. Chem. 2010, 2010, 999–1015. [Google Scholar] [CrossRef]
- Kim, Y.; Park, J.; Kim, M.-J. Dynamic Kinetic Resolution of Amines and Amino Acids by Enzyme-Metal Cocatalysis. ChemCatChem 2011, 3, 271–277. [Google Scholar] [CrossRef]
- Kourist, R.; Bornscheuer, U.T. Biocatalytic synthesis of optically active tertiary alcohols. Appl. Microbiol. Biotechnol. 2011, 91, 505–517. [Google Scholar] [CrossRef]
- Weiser, D.; Nagy, F.; Bánóczi, G.; Oláh, M.; Farkas, A.; Szilágyi, A.; László, K.; Gellért, Á.; Marosi, G.; Kemény, S.; et al. Immobilization engineering – How to design advanced sol–gel systems for biocatalysis? Green Chem. 2017, 19, 3927–3937. [Google Scholar] [CrossRef]
- Farkas, E.; Oláh, M.; Földi, A.; Kóti, J.; Éles, J.; Nagy, J.; Gal, C.A.; Paizs, C.; Hornyánszky, G.; Poppe, L. Chemoenzymatic Dynamic Kinetic Resolution of Amines in Fully Continuous-Flow Mode. Org. Lett. 2018, 20, 8052–8056. [Google Scholar] [CrossRef]
- Xing, X.; Jia, J.-Q.; Zhang, J.-F.; Zhou, Z.-W.; Li, J.; Wang, N.; Yu, X.-Q. CALB Immobilized onto Magnetic Nanoparticles for Efficient Kinetic Resolution of Racemic Secondary Alcohols: Long-Term Stability and Reusability. Molecules 2019, 24, 490. [Google Scholar] [CrossRef]
- Oláh, M.; Suba, S.; Boros, Z.; Kovács, P.; Gosselin, M.; Gaudreault, C.; Hornyánszky, G. Lipase B from Candida antarctica Immobilized on Epoxy-functionalized Hollow Silica Microspheres: Efficient Biocatalysts for Enantiomer Selective Acylation of Alcohols and Amines. Period. Polytech. Chem. Eng. 2018, 62, 519–532. [Google Scholar] [CrossRef]
- Moustafa, G.A.I.; Kasama, K.; Higashio, K.; Akai, S. Base-promoted lipase-catalyzed kinetic resolution of atropisomeric 1,1′-biaryl-2,2′-diols. RSC Adv. 2019, 9, 1165–1175. [Google Scholar] [CrossRef]
- Riva, S. Laccases: Blue enzymes for green chemistry. Trends Biotechnol. 2006, 24, 219–226. [Google Scholar] [CrossRef] [PubMed]
- Sokic-Lazic, D.; Arechederra, R.L.; Treu, B.L.; Minteer, S.D. Oxidation of Biofuels: Fuel Diversity and Effectiveness of Fuel Oxidation through Multiple Enzyme Cascades. Electroanalysis 2010, 22, 757–764. [Google Scholar] [CrossRef]
- Mitsou, E.; Xenakis, A.; Zoumpanioti, M. Oxidation Catalysis by Enzymes in Microemulsions. Catalysts 2017, 7, 52. [Google Scholar] [CrossRef]
- Hollmann, F.; Arends, I.W.C.E.; Holtmann, D. Enzymatic reductions for the chemist. Green Chem. 2011, 13, 2285–2313. [Google Scholar] [CrossRef]
- Durchschein, K.; Hall, M.; Faber, K. Unusual reactions mediated by FMN-dependent ene- and nitro-reductases. Green Chem. 2013, 15, 1764–1772. [Google Scholar] [CrossRef]
- Schrittwieser, J.H.; Velikogne, S.; Kroutil, W. Biocatalytic Imine Reduction and Reductive Amination of Ketones. Adv. Synth. Catal. 2015, 357, 1655–1685. [Google Scholar] [CrossRef]
- Bódai, V.; Nagy-Győr, L.; Örkényi, R.; Molnár, Z.; Kohári, S.; Erdélyi, B.; Nagymáté, Z.; Romsics, C.; Paizs, C.; Poppe, L.; et al. Wickerhamomyces subpelliculosus as whole-cell biocatalyst for stereoselective bioreduction of ketones. J. Mol. Catal. B Enzym. 2016, 134, 206–214. [Google Scholar] [CrossRef]
- Fuchs, M.; Farnberger, J.E.; Kroutil, W. The Industrial Age of Biocatalytic Transamination. European J. Org. Chem. 2015, 2015, 6965–6982. [Google Scholar] [CrossRef]
- Liu, J.; Wu, S.; Li, Z. Recent advances in enzymatic oxidation of alcohols. Curr. Opin. Chem. Biol. 2018, 43, 77–86. [Google Scholar] [CrossRef] [PubMed]
- Nagy-Győr, L.; Abaházi, E.; Bódai, V.; Sátorhelyi, P.; Erdélyi, B.; Balogh-Weiser, D.; Paizs, C.; Hornyánszky, G.; Poppe, L. Co-immobilized Whole Cells with ω-Transaminase and Ketoreductase Activities for Continuous-Flow Cascade Reactions. ChemBioChem 2018, 19, 1845–1848. [Google Scholar] [CrossRef] [PubMed]
- Guo, F.; Berglund, P. Transaminase biocatalysis: Optimization and application. Green Chem. 2017, 19, 333–360. [Google Scholar] [CrossRef] [Green Version]
- D. Patil, M.; Grogan, G.; Bommarius, A.; Yun, H. Recent Advances in ω-Transaminase-Mediated Biocatalysis for the Enantioselective Synthesis of Chiral Amines. Catalysts 2018, 8, 254. [Google Scholar] [CrossRef] [Green Version]
- Lenz, M.; Borlinghaus, N.; Weinmann, L.; Nestl, B.M. Recent advances in imine reductase-catalyzed reactions. World J. Microbiol. Biotechnol. 2017, 33, 199. [Google Scholar] [CrossRef]
- Liu, J.; Li, Z. Enhancing cofactor recycling in the bioconversion of racemic alcohols to chiral amines with alcohol dehydrogenase and amine dehydrogenase by coupling cells and cell-free system. Biotechnol. Bioeng. 2019, 116, 536–542. [Google Scholar] [CrossRef]
- Varga, A.; Bánóczi, G.; Nagy, B.; Bencze, L.C.; Toşa, M.I.; Gellért, Á.; Irimie, F.D.; Rétey, J.; Poppe, L.; Paizs, C. Influence of the aromatic moiety in α- and β-arylalanines on their biotransformation with phenylalanine 2,3-aminomutase from Pantoea agglomerans. RSC Adv. 2016, 6, 56412–56420. [Google Scholar] [CrossRef] [Green Version]
- Xue, Y.-P.; Cao, C.-H.; Zheng, Y.-G. Enzymatic asymmetric synthesis of chiral amino acids. Chem. Soc. Rev. 2018, 47, 1516–1561. [Google Scholar] [CrossRef]
- Gao, X.; Ma, Q.; Zhu, H. Distribution, industrial applications, and enzymatic synthesis of d -amino acids. Appl. Microbiol. Biotechnol. 2015, 99, 3341–3349. [Google Scholar] [CrossRef]
- Rosenthal, K.; Lütz, S. Recent developments and challenges of biocatalytic processes in the pharmaceutical industry. Curr. Opin. Green Sustain. Chem. 2018, 11, 58–64. [Google Scholar] [CrossRef]
- Kristinsson, H.G.; Rasco, B.A. Fish Protein Hydrolysates: Production, Biochemical, and Functional Properties. Crit. Rev. Food Sci. Nutr. 2000, 40, 43–81. [Google Scholar] [CrossRef] [PubMed]
- Leroy, F.; De Vuyst, L. Lactic acid bacteria as functional starter cultures for the food fermentation industry. Trends Food Sci. Technol. 2004, 15, 67–78. [Google Scholar] [CrossRef]
- Amine, A.; Mohammadi, H.; Bourais, I.; Palleschi, G. Enzyme inhibition-based biosensors for food safety and environmental monitoring. Biosens. Bioelectron. 2006, 21, 1405–1423. [Google Scholar] [CrossRef] [PubMed]
- Czinkóczky, R.; Németh, Á. Investigations into Enzymatic Bioconversion to Form Rebaudioside A from Stevioside. Period. Polytech. Chem. Eng. 2018, 62, 396–402. [Google Scholar] [CrossRef]
- Araya, E.; Urrutia, P.; Romero, O.; Illanes, A.; Wilson, L. Design of combined crosslinked enzyme aggregates (combi-CLEAs) of β-galactosidase and glucose isomerase for the one-pot production of fructose syrup from lactose. Food Chem. 2019, 288, 102–107. [Google Scholar] [CrossRef] [PubMed]
- Straathof, A.J.; Panke, S.; Schmid, A. The production of fine chemicals by biotransformations. Curr. Opin. Biotechnol. 2002, 13, 548–556. [Google Scholar] [CrossRef]
- Panke, S.; Held, M.; Wubbolts, M. Trends and innovations in industrial biocatalysis for the production of fine chemicals. Curr. Opin. Biotechnol. 2004, 15, 272–279. [Google Scholar] [CrossRef]
- Thompson, M.P.; Peñafiel, I.; Cosgrove, S.C.; Turner, N.J. Biocatalysis Using Immobilized Enzymes in Continuous Flow for the Synthesis of Fine Chemicals. Org. Process Res. Dev. 2019, 23, 9–18. [Google Scholar] [CrossRef]
- Lee, L.J.; Yang, S.; Lai, S.; Bai, Y.; Huang, W.; Juang, Y. Microfluidic Enzyme-Linked Immunosorbent Assay Technology. In Advances in Clinical Chemistry; Elsevier: Amsterdam, The Netherlands, 2006; Volume 42, pp. 255–295. ISBN 0120103427. [Google Scholar]
- Haun, J.B.; Yoon, T.-J.; Lee, H.; Weissleder, R. Magnetic nanoparticle biosensors. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2010, 2, 291–304. [Google Scholar] [CrossRef]
- De la Rica, R.; Aili, D.; Stevens, M.M. Enzyme-responsive nanoparticles for drug release and diagnostics. Adv. Drug Deliv. Rev. 2012, 64, 967–978. [Google Scholar] [CrossRef]
- El Harrad, L.; Bourais, I.; Mohammadi, H.; Amine, A. Recent Advances in Electrochemical Biosensors Based on Enzyme Inhibition for Clinical and Pharmaceutical Applications. Sensors 2018, 18, 164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, H.H.; Lee, S.H.; Lee, U.J.; Fermin, C.D.; Kim, M. Immobilized Enzymes in Biosensor Applications. Materials 2019, 12, 121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dror, A.; Kanteev, M.; Kagan, I.; Gihaz, S.; Shahar, A.; Fishman, A. Structural insights into methanol-stable variants of lipase T6 from Geobacillus stearothermophilus. Appl. Microbiol. Biotechnol. 2015, 99, 9449–9461. [Google Scholar] [CrossRef] [PubMed]
- Gihaz, S.; Weiser, D.; Dror, A.; Sátorhelyi, P.; Jerabek-Willemsen, M.; Poppe, L.; Fishman, A. Creating an Efficient Methanol-Stable Biocatalyst by Protein and Immobilization Engineering Steps towards Efficient Biosynthesis of Biodiesel. ChemSusChem 2016, 9, 3161–3170. [Google Scholar] [CrossRef] [PubMed]
- Douka, A.; Vouyiouka, S.; Papaspyridi, L.-M.; Papaspyrides, C.D. A review on enzymatic polymerization to produce polycondensation polymers: The case of aliphatic polyesters, polyamides and polyesteramides. Prog. Polym. Sci. 2018, 79, 1–25. [Google Scholar] [CrossRef]
- Su, J.; Wang, C.; Noro, J.; Cavaco-Paulo, A.; Silva, C.; Fu, J. Polymers from Bamboo Extracts Produced by Laccase. Polymers 2018, 10, 1141. [Google Scholar] [CrossRef] [Green Version]
- Jiang, Y.; Loos, K. Enzymatic Synthesis of Biobased Polyesters and Polyamides. Polymers 2016, 8, 243. [Google Scholar] [CrossRef] [Green Version]
- Rokhati, N.; Susanto, H.; Haryani, K.; Pramudono, B. Enhanced Enzymatic Hydrolysis of Chitosan by Surfactant Addition. Period. Polytech. Chem. Eng. 2017, 62, 286–291. [Google Scholar] [CrossRef] [Green Version]
- Gong, J.; Kong, T.; Li, Y.; Li, Q.; Li, Z.; Zhang, J. Biodegradation of Microplastic Derived from Poly(ethylene terephthalate) with Bacterial Whole-Cell Biocatalysts. Polymers 2018, 10, 1326. [Google Scholar] [CrossRef] [Green Version]
- Blackwell, C.; Haernvall, K.; Guebitz, G.; Groombridge, M.; Gonzales, D.; Khosravi, E. Enzymatic Degradation of Star Poly(ε-Caprolactone) with Different Central Units. Polymers 2018, 10, 1266. [Google Scholar] [CrossRef] [Green Version]
- Poppe, L.; Vértessy, B.G. The Fourth Wave of Biocatalysis Emerges–The 13th International Symposium on Biocatalysis and Biotransformations. ChemBioChem 2018, 19, 284–287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amid, A.; Hassan, N.; Jamaluddin, M.J.A.; Othman, M.E.F.; Belgasem, F.F.B.; Salleh, H.M.; Yusof, F.; Ismail, N.A.; Azmi, A.S.; Sulaiman, S.; et al. Recombinant Enzymes—From Basic Science to Commercialization; Amid, A., Ed.; Springer International Publishing: Heilderberg, 2015; ISBN 978-3-319-12396-7. [Google Scholar]
- Young, C.L.; Britton, Z.T.; Robinson, A.S. Recombinant protein expression and purification: A comprehensive review of affinity tags and microbial applications. Biotechnol. J. 2012, 7, 620–634. [Google Scholar] [CrossRef] [PubMed]
- Terpe, K. Overview of tag protein fusions: From molecular and biochemical fundamentals to commercial systems. Appl. Microbiol. Biotechnol. 2003, 60, 523–533. [Google Scholar] [CrossRef] [PubMed]
- Rosano, G.L.; Ceccarelli, E.A. Recombinant protein expression in Escherichia coli: Advances and challenges. Front. Microbiol. 2014, 5, 1–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vahidi, A.K.; Yang, Y.; Ngo, T.P.N.; Li, Z. Simple and Efficient Immobilization of Extracellular His-Tagged Enzyme Directly from Cell Culture Supernatant As Active and Recyclable Nanobiocatalyst: High-Performance Production of Biodiesel from Waste Grease. ACS Catal. 2015, 5, 3157–3161. [Google Scholar] [CrossRef]
- Peternel, Š. Bacterial cell disruption: A crucial step in protein production. N. Biotechnol. 2013, 30, 250–254. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Ding, L.; Sun, J.; Boussetta, N.; Vorobiev, E. Yeast cell disruption strategies for recovery of intracellular bio-active compounds—A review. Innov. Food Sci. Emerg. Technol. 2016, 36, 181–192. [Google Scholar] [CrossRef]
- Shehadul Islam, M.; Aryasomayajula, A.; Selvaganapathy, P.R. A Review on Macroscale and Microscale Cell Lysis Methods. Micromachines 2017, 8, 83. [Google Scholar] [CrossRef]
- Hatti-Kaul, R.; Mattiasson, B. Isolation and purification of proteins; Hatti-Kaul, R., Mattiasson, B., Eds.; Marcel Dekker, Inc.: New York, NY, USA, 2012; ISBN 0824707265. [Google Scholar]
- Waugh, D.S. An overview of enzymatic reagents for the removal of affinity tags. Protein Expr. Purif. 2011, 80, 283–293. [Google Scholar] [CrossRef]
- Kranen, E.; Detzel, C.; Weber, T.; Jose, J. Autodisplay for the co-expression of lipase and foldase on the surface of E. coli: Washing with designer bugs. Microb. Cell Fact. 2014, 13, 19. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Jin, X.; Lu, X.; Chu, F.; Shen, J.; Ma, Y.; Liu, M.; Zhu, J. Construction and characterization of a thermostable whole-cell chitinolytic enzyme using yeast surface display. World J. Microbiol. Biotechnol. 2014, 30, 2577–2585. [Google Scholar] [CrossRef] [PubMed]
- Saraswat, M.; Musante, L.; Ravidá, A.; Shortt, B.; Byrne, B.; Holthofer, H. Preparative Purification of Recombinant Proteins: Current Status and Future Trends. Biomed Res. Int. 2013, 2013, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Stolarzewicz, I.; Białecka-Florjańczyk, E.; Majewska, E.; Krzyczkowska, J. Immobilization of Yeast on Polymeric Supports. Chem. Biochem. Eng. Q. 2011, 25, 135–144. [Google Scholar]
- Kisukuri, C.M.; Andrade, L.H. Production of chiral compounds using immobilized cells as a source of biocatalysts. Org. Biomol. Chem. 2015, 13, 10086–10107. [Google Scholar] [CrossRef]
- Melvik, J.E.; Dornish, M. Alginate as a Carrier for Cell Immobilisation. In Fundamentals of Cell Immobilisation Biotechnology; Nedović, V., Willaert, R., Eds.; Springer: Dordrecht, The Netherlands, 2004; pp. 33–51. ISBN 978-94-017-1638-3. [Google Scholar]
- Michelini, E.; Roda, A. Staying alive: New perspectives on cell immobilization for biosensing purposes. Anal. Bioanal. Chem. 2012, 402, 1785–1797. [Google Scholar] [CrossRef]
- Krajewska, B. Application of chitin- and chitosan-based materials for enzyme immobilizations: A review. Enzyme Microb. Technol. 2004, 35, 126–139. [Google Scholar] [CrossRef]
- Carballeira, J.D.; Quezada, M.A.; Hoyos, P.; Simeó, Y.; Hernaiz, M.J.; Alcantara, A.R.; Sinisterra, J.V. Microbial cells as catalysts for stereoselective red–ox reactions. Biotechnol. Adv. 2009, 27, 686–714. [Google Scholar] [CrossRef]
- Zajkoska, P.; Rebroš, M.; Rosenberg, M. Biocatalysis with immobilized Escherichia coli. Appl. Microbiol. Biotechnol. 2013, 97, 1441–1455. [Google Scholar] [CrossRef]
- Desimone, M.F.; Alvarez, G.S.; Foglia, M.L.; Diaz, L.E. Development of Sol-Gel Hybrid Materials for Whole Cell Immobilization. Recent Pat. Biotechnol. 2009, 3, 55–60. [Google Scholar] [CrossRef]
- Desimone, M.F.; De Marzi, M.C.; Copello, G.J.; Fernández, M.M.; Malchiodi, E.L.; Diaz, L.E. Efficient preservation in a silicon oxide matrix of Escherichia coli, producer of recombinant proteins. Appl. Microbiol. Biotechnol. 2005, 68, 747–752. [Google Scholar] [CrossRef]
- Silva, C.R.; Airoldi, C. Acid and Base Catalysts in the Hybrid Silica Sol–Gel Process. J. Colloid Interface Sci. 1997, 195, 381–387. [Google Scholar] [CrossRef] [PubMed]
- Kato, K.; Nakamura, H.; Nakanishi, K. Asymmetric bioreduction of acetophenones by Baker’s yeast and its cell-free extract encapsulated in sol–gel silica materials. Appl. Surf. Sci. 2014, 293, 312–317. [Google Scholar] [CrossRef]
- Klein, S.; Kuhn, J.; Avrahami, R.; Tarre, S.; Beliavski, M.; Green, M.; Zussman, E. Encapsulation of Bacterial Cells in Electrospun Microtubes. Biomacromolecules 2009, 10, 1751–1756. [Google Scholar] [CrossRef] [PubMed]
- Salalha, W.; Kuhn, J.; Dror, Y.; Zussman, E. Encapsulation of bacteria and viruses in electrospun nanofibres. Nanotechnology 2006, 17, 4675–4681. [Google Scholar] [CrossRef] [PubMed]
- Gensheimer, M.; Becker, M.; Brandis-Heep, A.; Wendorff, J.H.; Thauer, R.K.; Greiner, A. Novel Biohybrid Materials by Electrospinning: Nanofibers of Poly(ethylene oxide) and Living Bacteria. Adv. Mater. 2007, 19, 2480–2482. [Google Scholar] [CrossRef]
- Zussman, E. Encapsulation of cells within electrospun fibers. Polym. Adv. Technol. 2011, 22, 366–371. [Google Scholar] [CrossRef]
- Brosel-Oliu, S.; Mergel, O.; Uria, N.; Abramova, N.; van Rijn, P.; Bratov, A. 3D impedimetric sensors as a tool for monitoring bacterial response to antibiotics. Lab Chip 2019, 19, 1436–1447. [Google Scholar] [CrossRef] [Green Version]
- Jin, L.-Q.; Yang, B.; Xu, W.; Chen, X.-X.; Jia, D.-X.; Liu, Z.-Q.; Zheng, Y.-G. Immobilization of recombinant Escherichia coli whole cells harboring xylose reductase and glucose dehydrogenase for xylitol production from xylose mother liquor. Bioresour. Technol. 2019, 285, 121344. [Google Scholar] [CrossRef]
- Qiu, S.; Wang, Y.-J.; Yu, H.; Cheng, F.; Zheng, Y.-G. t-Butyl 6-cyano-(3R,5R)-dihydroxyhexanoate synthesis via asymmetric reduction by immobilized cells of carbonyl reductase and glucose dehydrogenase co-expression E. coli. Process Biochem. 2019, 80, 43–51. [Google Scholar] [CrossRef]
- Stojkovič, G.; Žnidaršič-Plazl, P. Continuous synthesis of l-malic acid using whole-cell microreactor. Process Biochem. 2012, 47, 1102–1107. [Google Scholar] [CrossRef]
- Zehnder, T.; Sarker, B.; Boccaccini, A.R.; Detsch, R. Evaluation of an alginate–gelatine crosslinked hydrogel for bioplotting. Biofabrication 2015, 7, 25001. [Google Scholar] [CrossRef] [PubMed]
- Thakur, A.; Panesar, P.S.; Saini, M.S. Parametric Optimization of Lactic Acid Production by Immobilized Lactobacillus casei Using Box-Behnken Design. Period. Polytech. Chem. Eng. 2018, 62, 274–285. [Google Scholar] [CrossRef]
- Mateo, C.; Palomo, J.M.; Fernandez-Lorente, G.; Guisan, J.M.; Fernandez-Lafuente, R. Improvement of enzyme activity, stability and selectivity via immobilization techniques. Enzyme Microb. Technol. 2007, 40, 1451–1463. [Google Scholar] [CrossRef]
- Taqieddin, E.; Amiji, M. Enzyme immobilization in novel alginate–chitosan core-shell microcapsules. Biomaterials 2004, 25, 1937–1945. [Google Scholar] [CrossRef]
- Sóti, P.L.; Weiser, D.; Vigh, T.; Nagy, Z.K.; Poppe, L.; Marosi, G. Electrospun polylactic acid and polyvinyl alcohol fibers as efficient and stable nanomaterials for immobilization of lipases. Bioprocess Biosyst. Eng. 2016, 39, 449–459. [Google Scholar] [CrossRef]
- Krisch, E.; Balogh-Weiser, D.; Klimko, J.; Gyarmati, B.; Laszlo, K.; Poppe, L.; Szilagyi, A. Composite beads of silica gel, alginate and poly(aspartic acid) for the immobilization of a lipase enzyme. Express Polym. Lett. 2019, 13, 512–523. [Google Scholar] [CrossRef]
- Teepoo, S.; Dawan, P.; Barnthip, N. Electrospun Chitosan-Gelatin Biopolymer Composite Nanofibers for Horseradish Peroxidase Immobilization in a Hydrogen Peroxide Biosensor. Biosensors 2017, 7, 47. [Google Scholar] [CrossRef] [Green Version]
- Jesionowski, T.; Zdarta, J.; Krajewska, B. Enzyme immobilization by adsorption: A review. Adsorption 2014, 20, 801–821. [Google Scholar] [CrossRef] [Green Version]
- Boros, Z.; Weiser, D.; Márkus, M.; Abaháziová, E.; Magyar, Á.; Tomin, A.; Koczka, B.; Kovács, P.; Poppe, L. Hydrophobic adsorption and covalent immobilization of Candida antarctica lipase B on mixed-function-grafted silica gel supports for continuous-flow biotransformations. Process Biochem. 2013, 48, 1039–1047. [Google Scholar] [CrossRef] [Green Version]
- Weiser, D.; Varga, A.; Kovács, K.; Nagy, F.; Szilágyi, A.; Vértessy, B.G.; Paizs, C.; Poppe, L. Bisepoxide Cross-Linked Enzyme Aggregates-New Immobilized Biocatalysts for Selective Biotransformations. ChemCatChem 2014, 6, 1463–1469. [Google Scholar] [CrossRef]
- Vazquez-Ortega, P.G.; Alcaraz-Fructuoso, M.T.; Rojas-Contreras, J.A.; López-Miranda, J.; Fernandez-Lafuente, R. Stabilization of dimeric β-glucosidase from Aspergillus niger via glutaraldehyde immobilization under different conditions. Enzyme Microb. Technol. 2018, 110, 38–45. [Google Scholar] [CrossRef] [PubMed]
- Turková, J.; Bláha, K.; Malaníková, M.; Vančurová, D.; Švec, F.; Kálal, J. Methacrylate gels with epoxide groups as supports for immobilization of enzymes in pH range 3-12. Biochim. Biophys. Acta 1978, 524, 162–169. [Google Scholar] [CrossRef]
- Abaházi, E.; Lestál, D.; Boros, Z.; Poppe, L. Tailoring the Spacer Arm for Covalent Immobilization of Candida antarctica Lipase B—Thermal Stabilization by Bisepoxide-Activated Aminoalkyl Resins in Continuous-Flow Reactors. Molecules 2016, 21, 767. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guisan, J.M.; López-Gallego, F.; Betancor, L.; Mateo, C.; Grazu, V.; Fernandez-Lorente, G.; Rocha-Martin, J.; Bolivar, J.M.; Ovsejevi, K.; Manta, C.; et al. Immobilization of Enzymes and Cells. In Methods in Molecular Biology, 3rd ed.; Guisan, J.M., Ed.; Humana Press: Totowa, NJ, USA, 2013; Volume 1051, ISBN 978-1-62703-549-1. [Google Scholar]
- Mateo, C.; Fernández-Lorente, G.; Abian, O.; Fernández-Lafuente, R.; Guisán, J.M. Multifunctional Epoxy Supports: A New Tool to Improve the Covalent Immobilization of Proteins. The Promotion of Physical Adsorptions of Proteins on the Supports before Their Covalent Linkage. Biomacromolecules 2000, 1, 739–745. [Google Scholar] [CrossRef]
- Mateo, C.; Bolivar, J.M.; Godoy, C.A.; Rocha-Martin, J.; Pessela, B.C.; Curiel, J.A.; Muñoz, R.; Guisan, J.M.; Fernández-Lorente, G. Improvement of Enzyme Properties with a Two-Step Immobilizaton Process on Novel Heterofunctional Supports. Biomacromolecules 2010, 11, 3112–3117. [Google Scholar] [CrossRef] [Green Version]
- Mateo, C.; Grazu, V.; Palomo, J.M.; Lopez-Gallego, F.; Fernandez-Lafuente, R.; Guisan, J.M. Immobilization of enzymes on heterofunctional epoxy supports. Nat. Protoc. 2007, 2, 1022–1033. [Google Scholar] [CrossRef]
- Barbosa, O.; Torres, R.; Ortiz, C.; Berenguer-Murcia, Á.; Rodrigues, R.C.; Fernandez-Lafuente, R. Heterofunctional Supports in Enzyme Immobilization: From Traditional Immobilization Protocols to Opportunities in Tuning Enzyme Properties. Biomacromolecules 2013, 14, 2433–2462. [Google Scholar] [CrossRef] [Green Version]
- Grazú, V.; Abian, O.; Mateo, C.; Batista-Viera, F.; Fernández-Lafuente, R.; Guisán, J.M. Novel bifunctional epoxy/thiol-reactive support to immobilize thiol containing proteins by the epoxy chemistry. Biomacromolecules 2003, 4, 1495–1501. [Google Scholar] [CrossRef]
- Grazú, V.; Abian, O.; Mateo, C.; Batista-Viera, F.; Fernández-Lafuente, R.; Guisán, J.M. Stabilization of enzymes by multipoint immobilization of thiolated proteins on new epoxy-thiol supports. Biotechnol. Bioeng. 2005, 90, 597–605. [Google Scholar] [CrossRef]
- Manoel, E.A.; dos Santos, J.C.S.; Freire, D.M.G.; Rueda, N.; Fernandez-Lafuente, R. Immobilization of lipases on hydrophobic supports involves the open form of the enzyme. Enzyme Microb. Technol. 2015, 71, 53–57. [Google Scholar] [CrossRef]
- Barbosa, O.; Ortiz, C.; Torres, R.; Fernandez-Lafuente, R. Effect of the immobilization protocol on the properties of lipase B from Candida antarctica in organic media: Enantiospecific production of atenolol acetate. J. Mol. Catal. B Enzym. 2011, 71, 124–132. [Google Scholar] [CrossRef]
- Chen, C.-I.; Chen, C.-W.; Huang, C.-W.; Liu, Y.-C. Simultaneous purification and immobilization of penicillin G acylase using bifunctional membrane. J. Memb. Sci. 2007, 298, 24–29. [Google Scholar] [CrossRef]
- Tural, B.; Tural, S.; Ertaş, E.; Yalınkılıç, İ.; Demir, A.S. Purification and covalent immobilization of benzaldehyde lyase with heterofunctional chelate-epoxy modified magnetic nanoparticles and its carboligation reactivity. J. Mol. Catal. B Enzym. 2013, 95, 41–47. [Google Scholar] [CrossRef]
- Alsafadi, D.; Paradisi, F. Covalent Immobilization of Alcohol Dehydrogenase (ADH2) from Haloferax volcanii: How to Maximize Activity and Optimize Performance of Halophilic Enzymes. Mol. Biotechnol. 2014, 56, 240–247. [Google Scholar] [CrossRef]
- Mateo, C.; Fernández-Lorente, G.; Cortés, E.; Garcia, J.L.; Fernández-Lafuente, R.; Guisan, J.M. One-step purification, covalent immobilization, and additional stabilization of poly-His-tagged proteins using novel heterofunctional chelate-epoxy supports. Biotechnol. Bioeng. 2001, 76, 269–276. [Google Scholar] [CrossRef]
- Weiser, D.; Bencze, L.C.; Bánóczi, G.; Ender, F.; Kiss, R.; Kókai, E.; Szilágyi, A.; Vértessy, B.G.; Farkas, Ö.; Paizs, C.; et al. Phenylalanine Ammonia-Lyase-Catalyzed Deamination of an Acyclic Amino Acid: Enzyme Mechanistic Studies Aided by a Novel Microreactor Filled with Magnetic Nanoparticles. ChemBioChem 2015, 16, 2283–2288. [Google Scholar] [CrossRef]
- Sathishkumar, P.; Kamala-Kannan, S.; Cho, M.; Kim, J.S.; Hadibarata, T.; Salim, M.R.; Oh, B.-T. Laccase immobilization on cellulose nanofiber: The catalytic efficiency and recyclic application for simulated dye effluent treatment. J. Mol. Catal. B Enzym. 2014, 100, 111–120. [Google Scholar] [CrossRef]
- Bartha-Vári, J.H.; Bencze, L.C.; Bell, E.; Poppe, L.; Katona, G.; Irimie, F.-D.; Paizs, C.; Toșa, M.I. Aminated Single-walled Carbon Nanotubes as Carrier for Covalent Immobilization of Phenylalanine Ammonia-lyase. Period. Polytech. Chem. Eng. 2017, 61, 59–66. [Google Scholar] [CrossRef] [Green Version]
- Barbosa, O.; Ortiz, C.; Berenguer-Murcia, Á.; Torres, R.; Rodrigues, R.C.; Fernandez-Lafuente, R. Strategies for the one-step immobilization–purification of enzymes as industrial biocatalysts. Biotechnol. Adv. 2015, 33, 435–456. [Google Scholar] [CrossRef] [Green Version]
- Porath, J.; Carlsson, J.; Olsson, I.; Belfrage, G. Metal chelate affinity chromatography, a new approach to protein fractionation. Nature 1975, 258, 598–599. [Google Scholar] [CrossRef]
- Hochuli, E.; Döbeli, H.; Schacher, A. New metal chelate adsorbent selective for proteins and peptides cotaining neighbouring histidine residues. J. Chromatogr. A 1987, 411, 177–184. [Google Scholar] [CrossRef]
- Porath, J.; Olin, B. Immobilized Metal Ion Affinity Adsorption and Immobilized Metal Ion Affinity Chromatography of Biomaterials. Serum Protein Affinities for Gel-Immobilized Iron and Nickel Ions. Biochemistry 1983, 22, 1621–1630. [Google Scholar] [CrossRef] [PubMed]
- Sassenfeld, H.M.; Brewer, S.J. A polypeptide fusion designed for the purification of recombinant proteins. Bio/Technology 1984, 2, 76–81. [Google Scholar] [CrossRef]
- Hopp, T.P.; Prickett, K.S.; Price, V.L.; Libby, R.T.; March, C.J.; Cerretti, D.P.; Urdal, D.L.; Conlon, P.J. A short polypeptide marker sequence useful for recombinant protein identification and purification. Bio/Technology 1988, 6, 1204–1210. [Google Scholar] [CrossRef]
- Korndorfer, I.P.; Skerra, A. Improved affinity of engineered streptavidin for the Strep-tag II peptide is due to a fixed open conformation of the lid-like loop at the binding site. Protein Sci. 2002, 11, 883–893. [Google Scholar] [CrossRef]
- Stofko-Hahn, R.E.; Carr, D.W.; Scott, J.D. A single step purification for recombinant proteins Characterization of a microtubule associated protein (MAP 2) fragment which associates with the type II cAMP-dependent protein kinase. FEBS Lett. 1992, 302, 274–278. [Google Scholar] [CrossRef] [Green Version]
- Tomme, P.; Boraston, A.; McLean, B.; Kormos, J.; Creagh, A.L.; Sturch, K.; Gilkes, N.R.; Haynes, C.A.; Warren, R.A.J.; Kilburn, D.G. Characterization and affinity applications of cellulose-binding domains. J. Chromatogr. B Biomed. Sci. Appl. 1998, 715, 283–296. [Google Scholar] [CrossRef]
- Keefe, A.D.; Wilson, D.S.; Seelig, B.; Szostak, J.W. One-Step Purification of Recombinant Proteins Using a Nanomolar-Affinity Streptavidin-Binding Peptide, the SBP-Tag. Protein Expr. Purif. 2001, 23, 440–446. [Google Scholar] [CrossRef] [Green Version]
- Watanabe, T.; Ito, Y.; Yamada, T.; Hashimoto, M.; Sekine, S.; Tanaka, H. The roles of the C-terminal domain and type III domains of chitinase A1 from Bacillus circulans WL-12 in chitin degradation. J. Bacteriol. 1994, 176, 4465–4472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ikeda, T.; Ninomiya, K.-i.; Hirota, R.; Kuroda, A. Single-step affinity purification of recombinant proteins using the silica-binding Si-tag as a fusion partner. Protein Expr. Purif. 2010, 71, 91–95. [Google Scholar] [CrossRef]
- Bolivar, J.M.; Nidetzky, B. Positively charged mini-protein Z basic2 as a highly efficient silica binding module: Opportunities for enzyme immobilization on unmodified silica supports. Langmuir 2012, 28, 10040–10049. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.B.; Johnson, K.S. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene 1988, 67, 31–40. [Google Scholar] [CrossRef]
- Los, G.V.; Encell, L.P.; McDougall, M.G.; Hartzell, D.D.; Karassina, N.; Zimprich, C.; Wood, M.G.; Learish, R.; Ohana, R.F.; Urh, M.; et al. HaloTag: A Novel Protein Labeling Technology for Cell Imaging and Protein Analysis. ACS Chem. Biol. 2008, 3, 373–382. [Google Scholar] [CrossRef] [PubMed]
- Duplay, P.; Hofnung, M. Two regions of mature periplasmic maltose-binding protein of Escherichia coli involved in secretion. J. Bacteriol. 1988, 170, 4445–4450. [Google Scholar] [CrossRef] [Green Version]
- Hernandez, K.; Fernandez-Lafuente, R. Control of protein immobilization: Coupling immobilization and site-directed mutagenesis to improve biocatalyst or biosensor performance. Enzyme Microb. Technol. 2011, 48, 107–122. [Google Scholar] [CrossRef]
- Nagy, F.; Tasnádi, G.; Balogh-Weiser, D.; Bell, E.; Hall, M.; Faber, K.; Poppe, L. Smart Nanoparticles for Selective Immobilization of Acid Phosphatases. ChemCatChem 2018, 10, 3490–3499. [Google Scholar] [CrossRef] [Green Version]
- Paizs, C.; Katona, A.; Rétey, J. The Interaction of Heteroaryl-Acrylates and Alanines with Phenylalanine Ammonia-Lyase from Parsley. Chem. A Eur. J. 2006, 12, 2739–2744. [Google Scholar] [CrossRef]
- Gloge, A.; Zoń, J.; Kövári, Á.; Poppe, L.; Rétey, J. Phenylalanine Ammonia-Lyase: The Use of Its Broad Substrate Specificity for Mechanistic Investigations and Biocatalysis—Synthesis of l-Arylalanines. Chem. Eur. J. 2000, 6, 3386–3390. [Google Scholar] [CrossRef]
- Hydery, T.; Coppenrath, V.A. A Comprehensive Review of Pegvaliase, an Enzyme Substitution Therapy for the Treatment of Phenylketonuria. Drug Target Insights 2019, 13, 1–8. [Google Scholar] [CrossRef]
- Fields, C.; Li, P.; O’Mahony, J.J.; Lee, G.U. Advances in affinity ligand-functionalized nanomaterials for biomagnetic separation. Biotechnol. Bioeng. 2016, 113, 11–25. [Google Scholar] [CrossRef] [Green Version]
- Dima, N.A.; Filip, A.; Bencze, L.C.; Oláh, M.; Sátorhelyi, P.; Vértessy, B.G.; Poppe, L.; Paizs, C. Expression and purification of recombinant phenylalanine ammonia-lyase from Petroselinum crispum. Stud. Univ. Babes-Bolyai Chem. 2016, 61, 21–34. [Google Scholar]
- Lu, H.-T. Synthesis and characterization of amino-functionalized silica nanoparticles. Colloid J. 2013, 75, 311–318. [Google Scholar] [CrossRef]




| Tag Name | Length | Binding Matrix |
|---|---|---|
| Poly Arg-tag [128] | 5–6 arginine | Cation-exchange resin |
| Poly His-tag [126] | 6–10 histidine | Immobilized metal-coated support |
| FLAG [129] | 8 amino acids | Anti-FLAG MAbs |
| Strep-tag II [130] | 8 amino acids | Modified streptavidin |
| Calmodulin-binding peptide [131] | 26 amino acids | Calmodulin |
| Cellulose-binding domains [132] | 27–129 amino acids | Cellulose |
| SBP [133] | 38 amino acids | Streptavidin |
| Chitin-binding domain [134] | 51 amino acids | Chitin |
| Si-Tag (L2, Zbasic2 proteins) [135,136] | 58 and 273 amino acids | Silica surface |
| Glutathione S-transferase [137] | 211 amino acids | Glutathione |
| HaloTag [138] | 237 amino acids | HaloTag ligands |
| Maltose-binding protein [139] | 396 amino acids | Cross-linked amylose |
| PcPAL Solution | PcPAL-MNPs-NPDGE | PcPAL-MNPs-NPDGE/EDa-10 |
|---|---|---|
| Crude lysate | 0.4 ± 0.1 | 13.7 ± 0.6 |
| Purified PcPAL | 14.2 ± 0.5 | 14.1 ± 0.6 |
| Target Protein Concentration | PcPAL-MNPs-NPDGE | PcPAL-MNPs-NPDGE/EDa-10 | UBNPDGE/EDa−10/UBNPDGE |
|---|---|---|---|
| 5 | 0.5 | 12.3 | 24.1 |
| 10 | 1.0 | 12.6 | 13.2 |
| 25 | 3.3 | 13.2 | 4.0 |
| 50 | 8.3 | 13.4 | 1.6 |
| 75 | 11.9 | 13.6 | 1.2 |
| 90 | 12.4 | 13.6 | 1.1 |

| Reaction Cycle | PcPAL-MNPs-NPDGE/EDa-10 | PcPAL-MNPs-THPMTGE/EDa-10 | ||
|---|---|---|---|---|
| c [%] | eed-PHE [%] | c [%] | eed-PHE [%] | |
| 1 | 49 | 82 | 52 | 93 |
| 2 | 48 | 78 | 51 | 92 |
| 3 | 45 | 71 | 48 | 91 |
| 4 | 43 | 67 | 47 | 91 |
| 5 | 43 | 64 | 47 | 89 |

| Reaction cycle | PcPAL-MNPs-NPDGE/EDa-10 | PcPAL-MNPs-THPMTGE/EDa-10 | ||
|---|---|---|---|---|
| c [%] | eel-PHE [%] | c [%] | eel-PHE [%] | |
| 1 | 85 | »99 | 86 | »99 |
| 2 | 86 | »99 | 87 | »99 |
| 3 | 85 | »99 | 86 | »99 |
| 4 | 82 | »99 | 86 | »99 |
| 5 | 70 | »99 | 83 | »99 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sánta-Bell, E.; Molnár, Z.; Varga, A.; Nagy, F.; Hornyánszky, G.; Paizs, C.; Balogh-Weiser, D.; Poppe, L. “Fishing and Hunting”—Selective Immobilization of a Recombinant Phenylalanine Ammonia-Lyase from Fermentation Media. Molecules 2019, 24, 4146. https://doi.org/10.3390/molecules24224146
Sánta-Bell E, Molnár Z, Varga A, Nagy F, Hornyánszky G, Paizs C, Balogh-Weiser D, Poppe L. “Fishing and Hunting”—Selective Immobilization of a Recombinant Phenylalanine Ammonia-Lyase from Fermentation Media. Molecules. 2019; 24(22):4146. https://doi.org/10.3390/molecules24224146
Chicago/Turabian StyleSánta-Bell, Evelin, Zsófia Molnár, Andrea Varga, Flóra Nagy, Gábor Hornyánszky, Csaba Paizs, Diána Balogh-Weiser, and László Poppe. 2019. "“Fishing and Hunting”—Selective Immobilization of a Recombinant Phenylalanine Ammonia-Lyase from Fermentation Media" Molecules 24, no. 22: 4146. https://doi.org/10.3390/molecules24224146
APA StyleSánta-Bell, E., Molnár, Z., Varga, A., Nagy, F., Hornyánszky, G., Paizs, C., Balogh-Weiser, D., & Poppe, L. (2019). “Fishing and Hunting”—Selective Immobilization of a Recombinant Phenylalanine Ammonia-Lyase from Fermentation Media. Molecules, 24(22), 4146. https://doi.org/10.3390/molecules24224146