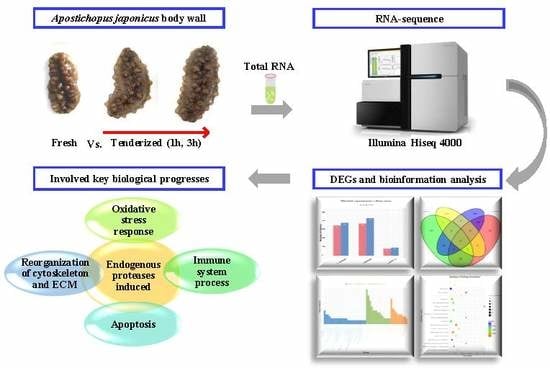
RNA Sequencing Analysis to Capture the Transcriptome Landscape during Tenderization in Sea Cucumber Apostichopus japonicus
Abstract
:1. Introduction
2. Results
2.1. Transcriptome Sequencing and Quality Control
2.2. Reads Mapping to the Reference Genome Dataset
2.3. Analysis of DEGs
2.4. Validation of the Results by RT-qPCR
2.5. Gene Ontology Analysis of DEGs
2.6. Pathway Enrichment Analysis of DEGs
3. Discussion
3.1. Genes of Endogenous Protease
3.2. Genes Associated with Oxidative Stress Response
3.3. Genes Associated with Immune Response
3.4. Genes Associated with Apoptosis Process
3.5. Genes Associated with Reorganization of Cytoskeleton and ECM
4. Materials and Methods
4.1. Animals Materials
4.2. mRNA Library Construction and Sequencing
4.3. RNA-seq Reads Mapping and DEG Testing
4.4. RT-qPCR Validation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| AJBW | Apostichopus japonicus body wall |
| Cyt b | Cytochrome b |
| DEGs | differential expression genes |
| ECM | extracellular matrix |
| ER | endoplasmic reticulum |
| FPKMs | Fragments Per Kilobase of exon model per Million mapped reads |
| FREPs | fibrinogen-related proteins |
| Gb | giga bases |
| GMP | gelatinolytic metalloproteinase |
| GO | Gene Ontology |
| GPx | glutathione peroxidase |
| GST | glutathione S-transferase |
| HIF-1 | hypoxia inducible factor-1 |
| HISAT | Hierarchical Indexing for Spliced Alignment of Transcripts |
| HSPs | heat shock proteins |
| IL-1 | interleukin-1 |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| LBLLPS | lactose-binding lectinslipopolysaccharide |
| MAC | membrane attack complex |
| MASP | mannan-binding lectin serine protease |
| MBL | mannose-binding lectin |
| MMP | matrix metalloproteinase |
| Nr | nonredundant protein sequences |
| PUF60 | poly-U-binding factor 60 kDa |
| RT-qPCR | quantitative Real-time Polymerase Chain Reaction |
| Rap-1 | Ras-proximate-1 |
| ROS | reactive oxygen species |
| TGF-β1 | transforming growth factor-1 |
| TNF | tumor necrosis factor |
References
- Wu, H.T.; Li, D.M.; Zhu, B.W.; Sun, J.J.; Zheng, J.; Wang, F.L.; Konno, K.; Jiang, X. Proteolysis of noncollagenous proteins in sea cucumber, Stichopus japonicus, body wall: Characterisation and the effects of cysteine protease inhibitors. Food Chem. 2013, 141, 1287–1294. [Google Scholar] [CrossRef] [PubMed]
- Bureau of Fisheries in Ministry of Agriculture. China Fishery Statistical Yearbook; China Agriculture Press: Beijing, China, 2017; p. 23. [Google Scholar]
- Qi, H.; Fu, H.; Dong, X.; Feng, D.; Li, N.; Wen, C.; Nakamura, Y.; Zhu, B. Apoptosis induction is involved in UVA-induced autolysis in sea cucumber Stichopus japonicus. J. Photochem. Photobiol. B 2016, 158, 130–135. [Google Scholar] [CrossRef] [PubMed]
- Qi, H.; Dong, X.F.; Zhao, Y.P.; Li, N.; Fu, H.; Feng, D.D.; Liu, L.; Yu, C.X. ROS production in homogenate from the body wall of sea cucumber Stichopus japonicus under UVA irradiation: ESR spin-trapping study. Food Chem. 2016, 192, 358–362. [Google Scholar] [CrossRef] [PubMed]
- Christensen, L.; Gunvig, A.; Torngren, M.A.; Aaslyng, M.D.; Knochel, S.; Christensen, M. Sensory characteristics of meat cooked for prolonged times at low temperature. Meat Sci. 2012, 90, 485–489. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Huang, F.; Xue, M.; Xu, X.; Zhou, G. The effect of active caspase-3 on degradation of chicken myofibrillar proteins and structure of myofibrils. Food Chem. 2011, 128, 22–27. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.P.; Li, Y.; Li, Y.; Song, L.; Cheng, S.S.; Li, D.M.; Zhu, B.W.; Zhou, D.Y.; Tan, M.Q. Combination of NMR and MRI techniques for non-invasive assessment of sea cucumber (Stichopus japonicas) tenderization during low-temperature heating process. Food Anal. Methods 2016. [Google Scholar] [CrossRef]
- Bi, J.R.; Li, Y.; Cheng, S.S.; Dong, X.P.; Kamal, T.; Zhou, D.Y.; Li, D.M.; Jiang, P.F.; Zhu, B.W.; Tan, M.Q. Changes in body wall of sea cucumber (Stichopus japonicus) during a two-step heating process assessed by rheology, LF-NMR, and texture profile analysis. Food Biophys. 2016, 11, 257–265. [Google Scholar] [CrossRef]
- Zhong, C.; Cai, Q.F.; Liu, G.M.; Sun, L.C.; Hara, K.; SU, W.J.; Cao, M.J. Purification and characterisation of cathepsin L from the skeletal muscle of blue scad (Decapterus maruadsi) and comparison of its role with myofibril-bound serine proteinase in the degradation of myofibrillar proteins. Food Chem. 2012, 133, 1560–1568. [Google Scholar] [CrossRef]
- Wang, K.K. Calpain and caspase: Can you tell the difference? Trends Neurosci. 2000, 23, 20–26. [Google Scholar] [CrossRef]
- Chen, L.; Feng, X.C.; Zhang, Y.Y.; Liu, X.B.; Zhang, W.G.; Li, C.B.; Ullah, N.; Xu, X.L.; Zhou, G.H. Effects of ultrasonic processing on caspase-3, calpain expression and myofibrillar structure of chicken during post-mortem ageing. Food Chem. 2015, 177, 280–287. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.P.; Zhu, B.W.; Sun, L.M.; Zheng, J.; Jiang, D.; Zhou, D.Y.; Wu, H.T.; Murata, Y. Changes of collagen in sea cucumber (Stichopus japonicas) during cooking. Food Sci. Biotechnol. 2011, 20, 1137–1141. [Google Scholar] [CrossRef]
- Qin, L.; Bi, J.R.; Li, D.M.; Dong, M.; Zhao, Z.Y.; Dong, X.P.; Zhou, D.Y.; Zhu, B.W. Unfolding/refolding study on collagen from sea cucumber based on 2D fourier transform infrared spectroscopy. Molecules 2016, 21, 1546. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.; Huang, M.; Zhang, H.; Zhang, C.; Zhang, D.; Zhou, G. Changes in apoptotic factors and caspase activation pathways during the postmortem aging of beef muscle. Food Chem. 2016, 190, 110–114. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.L.; Han, L.; Ma, X.L.; Yu, Q.L.; Zhao, S.N. Effect of mitochondrial apoptotic activation through the mitochondrial membrane permeability transition pore on yak meat tenderness during postmortem aging. Food Chem. 2017, 234, 323–331. [Google Scholar] [CrossRef] [PubMed]
- Tan, M.H.; Au, K.F.; Yablonovitch, A.L.; Wills, A.E.; Chuang, J.; Baker, J.C.; Wong, W.H.; Li, J.B. RNA sequencing reveals a diverse and dynamic repertoire of the Xenopus tropicalis transcriptome over development. Genome Res. 2013, 23, 201–216. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Chen, M.; Yang, H.; Wang, T.; Liu, B.; Shu, C.; Gardiner, D.M. Large scale gene expression profiling during intestine and body wall regeneration in the sea cucumber Apostichopus japonicus. Comp. Biochem. Physiol. Part D Genom. Proteom. 2011, 6, 195–205. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Feng, W.; Qiu, L.; Xia, C.; Su, X.; Jin, C.; Zhou, T.; Zeng, Y.; Li, T. Characterization of skin ulceration syndrome associated microRNAs in sea cucumber Apostichopus japonicus by deep sequencing. Fish Shellfish Immunol. 2012, 33, 436–441. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Yang, H.; Storey, K.B.; Chen, M. Differential gene expression in the respiratory tree of the sea cucumber Apostichopus japonicus during aestivation. Mar. Genom. 2014, 18, 173–183. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.C.; Dong, Y.; Sun, H.J.; Yang, A.F.; Chen, Z.; Gao, S.; Jiang, J.W.; Guan, X.Y.; Jiang, B.; Wang, B. Transcriptome sequencing of sea cucumber (Apostichopus japonicus) and the identification of gene-associated markers. Mol. Ecol. Resour. 2014, 14, 127–138. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Huang, J.; Liu, Z.; Zhou, Y.; Xia, B.; Wang, Y.; Kang, Y.; Wang, J. Transcriptome analysis provides insights into hepatic responses to moderate heat stress in the rainbow trout (Oncorhynchus mykiss). Gene 2017, 619, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Shiel, B.P.; Hall, N.E.; Cooke, I.R.; Robinson, N.A.; Strugnell, J.M. De novo characterisation of the greenlip abalone transcriptome (Haliotis laevigata) with a focus on the heat shock protein 70 (HSP70) family. Mar. Biotechnol. 2015, 17, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Barshis, D.J.; Ladner, J.T.; Oliver, T.A.; Palumbi, S.R. Lineage-specific transcriptional profiles of Symbiodinium spp. unaltered by heat stress in a coral host. Mol. Biol. Evol. 2014, 31, 1343–1352. [Google Scholar] [CrossRef] [PubMed]
- Smolina, I.; Kollias, S.; Møller, E.F.; Lindeque, P.; Sundaram, A.Y.M.; Ferandes, J.M.O.; Hoarau, G. Contrasting transcriptome response to thermal stress in two key zooplankton species, Calanus finmarchicus and C. glacialis. Mar. Ecol. Prog. Ser. 2015, 534, 79–95. [Google Scholar] [CrossRef]
- Liu, Y.X.; Zhou, D.Y.; Ma, D.D.; Liu, Z.Q.; Liu, Y.F.; Song, L.; Dong, X.P.; Li, D.M.; Zhu, B.W.; Konno, K.; et al. Effects of endogenous cysteine proteinases on structures of collagen fibres from dermis of sea cucumber (Stichopus japonicus). Food Chem. 2017, 232, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.M.; Wang, T.T.; Zhu, B.W.; Niu, H.L.; Zhang, R.; Hou, H.M.; Zhang, G.L.; Murata, Y. Effect of matrix metalloproteinase on autolysis of sea cucumber Stichopus japonicus. Food Sci. Biotechnol. 2013, 22, 1259–1261. [Google Scholar] [CrossRef]
- Zitka, O.; Kukacka, J.; Krizkova, S.; Huska, D.; Adam, V.; Masarik, M.; Prusa, R.; Kizek, R. Matrix metalloproteinases. Curr. Med. Chem. 2010, 17, 3751–3768. [Google Scholar] [CrossRef]
- The Sea Cucumber Apostichopus japonicus Genome. Available online: http://www.genedatabase.cn/aja_genome_20161129.html (accessed on 8 January 2018).
- Zhang, L.B.; Feng, Q.M.; Sun, L.N.; Fang, Y.; Xu, D.X.; Zhang, T.; Yang, H.S. Differential gene expression in the body wall of the sea cucumber (Apostichopus japonicus) under strong lighting and dark conditions. Acta Oceanol. Sin. 2018, 37. [Google Scholar] [CrossRef]
- Zhu, B.W.; Zhao, L.L.; Sun, L.M.; Li, D.M.; Murata, Y.; Yu, L.; Zhang, L. Purification and characterization of a cathepsin L-like enzyme from the body wall of the sea cucumber Stichopus japonicus. Biosci. Biotechnol. Biochem. 2014, 72, 1430–1437. [Google Scholar] [CrossRef]
- Zhong, M.; Hu, C.; Ren, C.; Luo, X.; Cai, Y. Characterization of a main extracellular matrix autoenzyme from the dermis of sea cucumber Stichopus monotuberculatus: Collagenase. Int. J. Food Prop. 2015, 19, 2495–2509. [Google Scholar] [CrossRef]
- Wu, H.L.; Hu, Y.Q.; Shen, J.D.; Cai, Q.F.; Liu, G.M.; Su, W.J.; Cao, M.J. Identification of a novel gelatinolytic metalloproteinase (GMP) in the body wall of sea cucumber (Stichopus japonicus) and its involvement in collagen degradation. Process Biochem. 2013, 48, 871–877. [Google Scholar] [CrossRef]
- Szlugit, G. The echinoderm collagen fibril: A hero in the connective tissue research of the 1990s. Bioessays 2007, 29, 645–653. [Google Scholar] [CrossRef] [PubMed]
- Saito, M.; Kunisaki, N.; Urano, N.; Kimura, S. Collagen as the major edible component of sea cucumber (Stichopus japonicus). J. Food Sci. 2002, 67, 1319–1322. [Google Scholar] [CrossRef]
- Mizuta, S.; Koizumi, Y.; Inoue, S.; Someya, C.; Hosoi, M.; Yokoyama, Y.; Yoshinaka, R. Existence of a 400kDa glycoprotein in the dermis of sea cucumber Apostichopus armata: Partial purification and characterization. Fish Sci. 2013, 79, 833–839. [Google Scholar] [CrossRef]
- Tallant, C.; Nodarse, A.M.; Gomis-Rüth, F.X. Matrix metalloproteinases: Fold and function of their catalytic domains. Biochim. Biophys. Acta Mol. Cell Res. 2009, 1803, 20–28. [Google Scholar] [CrossRef] [PubMed]
- Barshis, D.J.; Ladner, J.T.; Oliver, T.A.; Seneca, F.O.; Traylor-Knowles, N.; Palumbi, S.R. Genomic basis for coral resilience to climate change. Proc. Natl. Acad. Sci. USA 2013, 110, 1387–1392. [Google Scholar] [CrossRef] [PubMed]
- Lim, H.J.; Kim, B.M.; Hwang, I.J.; Lee, J.S.; Choi, I.Y.; Kim, Y.J.; Rhee, J.S. Thermal stress induces a distinct transcriptome profile in the Pacific oyster Crassostrea gigas. Comp. Biochem. Physiol. Part D Genom. Proteom. 2016, 19, 62–70. [Google Scholar] [CrossRef] [PubMed]
- Gleason, L.U.; Burton, R.S. RNA-seq reveals regional differences in transcriptome response to heat stress in the marine snail Chlorostoma funebralis. Mol. Ecol. 2015, 24, 610–627. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.F.; Qi, H.; Feng, D.D.; He, B.Y.; Nakamura, Y.; Yu, C.X.; Zhu, B.W. Oxidative stress involved in textural changes of sea cucumber Stichopus japonicus body wall (SJBW) during low-temperature treatment. Int. J. Food Prop. 2018, 21, 2646–2659. [Google Scholar] [CrossRef]
- Lang, R.P.; Bayne, C.J.; Camara, M.D.; Cunningham, C.; Jenny, M.J.; Langdon, C.J. Transcriptome profiling of selectively bred Pacific oyster Crassostrea gigas families that differ in tolerance of heat shock. Mar. Biotechnol. 2009, 11, 650–668. [Google Scholar] [CrossRef]
- Meistertzheim, A.L.; Tanguy, A.; Moraga, D.; Thebault, M.T. Identification of differentially expressed genes of the Pacific oyster Crassostrea gigas exposed to prolonged thermal stress. FEBS J. 2007, 274, 6392–6402. [Google Scholar] [CrossRef]
- Gullian Klanian, M.; Terrats Preciat, M. Effect of pH on temperature controlled degradation of reactive oxygen species, heat shock protein expression, and mucosal immunity in the sea cucumber Isostichopus badionotus. PLoS ONE 2017, 12, e0175812. [Google Scholar] [CrossRef] [PubMed]
- Mates, J.M. Effects of antioxidant enzymes in the molecular control of reactive oxygen species toxicology. Toxicol 2000, 153, 83–104. [Google Scholar] [CrossRef]
- Chen, Y.; Azad, M.B.; Gibson, S.B. Superoxide is the major reactive oxygen species regulating autophagy. Cell Death Differ. 2009, 16, 1040–1052. [Google Scholar] [CrossRef]
- Habte-Tsion, H.M.; Ren, M.; Liu, B.; Ge, X.; Xie, J.; Chen, R. Threonine modulates immune response, antioxidant status and gene expressions of antioxidant enzymes and antioxidant-immune-cytokine-related signaling molecules in juvenile blunt snout bream (Megalobrama amblycephala). Fish Shellfish Immunol. 2016, 51, 189–199. [Google Scholar] [CrossRef]
- Siwik, D.A.; Pagano, P.J.; Colucci, W.S. Oxidative stress regulates collagen synthesis and matrix metalloproteinase activity in cardiac fibroblasts. Am. J. Physiol. Cell Physiol. 2001, 280, C53–60. [Google Scholar] [CrossRef]
- Hoffmann, J.A.; Kafatos, F.C.; Janeway, C.A.; Ezekowitz, R.A. Phylogenetic perspectives in innate immunity. Science 1999, 284, 1313–1318. [Google Scholar] [CrossRef]
- Parks, W.C.; Wilson, C.L.; Lopez-Boado, Y.S. Matrix metalloproteinases as modulators of inflammation and innate immunity. Nat. Rev. Immunol. 2004, 4, 617–629. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Gomez, F.; Ortiz-Pineda, P.A.; Rivera-Cardona, G.; Garcia-Arraras, J.E. LPS-induced genes in intestinal tissue of the sea cucumber Holothuria glaberrima. PLoS ONE 2009, 4, e6178. [Google Scholar] [CrossRef]
- Zhang, X.; Sun, L.; Yuan, J.; Sun, Y.; Gao, Y.; Zhang, L.; Li, S.; Dai, H.; Hamel, J.F.; Liu, C.; et al. The sea cucumber genome provides insights into morphological evolution and visceral regeneration. PLoS Biol. 2017, 15, e2003790. [Google Scholar] [CrossRef]
- Wu, Y.; Yao, F.; Mei, Y.; Chu, B.; Cheng, C.; Liu, Y.; Li, X.; Zou, X.; Hou, L. Cloning and expression analysis of the gene encoding fibrinogen-like protein A, a novel regeneration-related protein from Apostichopus japonicus. Mol. Biol. Rep. 2014, 41, 2617–2627. [Google Scholar] [CrossRef]
- Soonthornchai, W.; Chaiyapechara, S.; Klinbunga, S.; Thongda, W.; Tangphatsornruang, S.; Yoocha, T.; Jarayabhand, P.; Jiravanichpaisal, P. Differentially expressed transcripts in stomach of Penaeus monodon in response to AHPND infection. Dev. Comp. Immunol. 2016, 65, 53–63. [Google Scholar] [CrossRef]
- Tanguy, M.; Gauthier-Clerc, S.; Pellerin, J.; Danger, J.M.; Siah, A. The immune response of Mytilus edulis hemocytes exposed to Vibrio splendidus LGP32 strain: A transcriptomic attempt at identifying molecular actors. Fish Shellfish Immunol. 2018, 74, 268–280. [Google Scholar] [CrossRef] [PubMed]
- Prasopdee, S.; Sotillo, J.; Tesana, S.; Laha, T.; Kulsantiwong, J.; Nolan, M.J.; Loukas, A.; Cantacessi, C. RNA-Seq reveals infection-induced gene expression changes in the snail intermediate host of the carcinogenic liver fluke, Opisthorchis viverrini. PLoS Negl. Trop. Dis. 2014, 8, e2765. [Google Scholar] [CrossRef] [PubMed]
- Mastellos, D.; Lambris, J.D. Complement: More than a ‘guard’ against invading pathogens? Trends Immunol. 2002, 23, 485–491. [Google Scholar] [CrossRef]
- Volanakis, J.E. Participation of C3 and its ligands in complement activation. Curr. Top. Microbiol. Immunol. 1990, 153, 1–21. [Google Scholar]
- Dong, Y.; Sun, H.; Zhou, Z.; Yang, A.; Chen, Z.; Guan, X.; Gao, S.; Wang, B.; Jiang, B.; Jiang, J. Expression analysis of immune related genes identified from the coelomocytes of sea cucumber (Apostichopus japonicus) in response to LPS challenge. Int. J. Mol. Sci. 2014, 15, 19472–19486. [Google Scholar] [CrossRef]
- Yang, A.; Zhou, Z.; Pan, Y.; Jiang, J.; Dong, Y.; Guan, X.; Sun, H.; Gao, S.; Chen, Z. RNA sequencing analysis to capture the transcriptome landscape during skin ulceration syndrome progression in sea cucumber Apostichopus japonicus. BMC Genom. 2016, 17, 459. [Google Scholar] [CrossRef]
- Nauta, A.J.; Daha, M.R.; Tijsma, O.; van de Water, B.; Tedesco, F.; Roos, A. The membrane attack complex of complement induces caspase activation and apoptosis. Eur. J. Immunol. 2002, 32, 783–792. [Google Scholar] [CrossRef]
- Yang, A.; Zhou, Z.; Dong, Y.; Jiang, B.; Wang, X.; Chen, Z.; Guan, X.; Wang, B.; Sun, D. Expression of immune-related genes in embryos and larvae of sea cucumber Apostichopus japonicus. Fish Shellfish Immunol. 2010, 29, 839–845. [Google Scholar] [CrossRef]
- Barat, A.; Sahoo, P.K.; Kumar, R.; Goel, C.; Singh, A.K. Transcriptional response to heat shock in liver of snow trout (Schizothorax richardsonii)—A vulnerable Himalayan Cyprinid fish. Funct. Integr. Genom. 2016, 16, 203–213. [Google Scholar] [CrossRef]
- Zhang, L.B.; Feng, Q.M.; SUn, L.N.; Ding, K.; Huo, D.; Yan, F.; Zhang, T.; Yang, H.S. Differential gene expression in the intestine of sea cucumber (Apostichopus japonicus) under low and high salinity conditions. Comp. Biochem. Physiol. Part D Genom. Proteom. 2018. [Google Scholar] [CrossRef]
- Cao, Y.; Ohwatari, N.; Matsumoto, T.; Kosaka, M.; Ohtsuru, A.; Yamashita, S. TGF-beta1 mediates 70-kDa heat shock protein induction due to ultraviolet irradiation in human skin fibroblasts. Eur. J. Physiol. 1999, 438, 239–244. [Google Scholar] [CrossRef]
- Shao, Y.; Li, C.; Chen, X.; Zhang, P.; Li, Y.; Li, T.; Jiang, J. Metabolomic responses of sea cucumber Apostichopus japonicus to thermal stresses. Aquaculture 2015, 435, 390–397. [Google Scholar] [CrossRef]
- Roberts, R.J.; Agius, C.; Saliba, C.; Bossier, P.; Sung, Y.Y. Heat shock proteins (chaperones) in fish and shellfish and their potential role in relation to fish health: A review. J. Fish Dis. 2010, 33, 789–801. [Google Scholar] [CrossRef]
- Nelson, R.J.; Ziegelhoffer, T.; Nicolet, C.; Werner-Washburne, M.; Craig, E.A. The translation machinery and 70 kd heat shock protein cooperate in protein synthesis. Cell 1992, 71, 97–105. [Google Scholar] [CrossRef]
- Shen, L.; Song, Y.; Fu, Y.; Li, P. MiR-29b mimics promotes cell apoptosis of smooth muscle cells via targeting on MMP-2. Cytotechnol 2018, 70, 351–359. [Google Scholar] [CrossRef]
- Ren, C.; Chen, T.; Sun, H.; Jiang, X.; Hu, C.; Qian, J.; Wang, Y. The first echinoderm poly-U-binding factor 60 kDa (PUF60) from sea cucumber (Stichopus monotuberculatus): Molecular characterization, inducible expression and involvement of apoptosis. Fish Shellfish Immunol. 2015, 47, 196–204. [Google Scholar] [CrossRef]
- Lee, G.H.; Kim, H.K.; Chae, S.W.; Kim, D.S.; Ha, K.C.; Cuddy, M.; Kress, C.; Reed, J.C.; Kim, H.R.; Chae, H.J. Bax inhibitor-1 regulates endoplasmic reticulum stress-associated reactive oxygen species and heme oxygenase-1 expression. J. Biol. Chem. 2007, 282, 21618–21628. [Google Scholar] [CrossRef]
- Reinehr, R.; Becker, S.; Wettstein, M.; Haussinger, D. Involvement of the Src family kinase yes in bile salt-induced apoptosis. Gastroenterol 2004, 127, 1540–1557. [Google Scholar] [CrossRef]
- Liu, S.; Wang, X.; Sun, F.; Zhang, J.; Feng, J.; Liu, H.; Rajendran, K.V.; Sun, L.; Zhang, Y.; Jiang, Y.; et al. RNA-seq reveals expression signatures of genes involved in oxygen transport, protein synthesis, folding, and degradation in response to heat stress in catfish. Physiol. Genom. 2013, 45, 462–476. [Google Scholar] [CrossRef]
- Artigaud, S.; Richard, J.; Thorne, M.A.; Lavaud, R.; Flye-Sainte-Marie, J.; Jean, F.; Peck, L.S.; Clark, M.S.; Pichereau, V. Deciphering the molecular adaptation of the king scallop (Pecten maximus) to heat stress using transcriptomics and proteomics. BMC Genom. 2015, 16, 988–1001. [Google Scholar] [CrossRef] [PubMed]
- Polato, N.R.; Voolstra, C.R.; Schnetzer, J.; DeSalvo, M.K.; Randall, C.J.; Szmant, A.M.; Medina, M.; Baums, I.B. Location-specific responses to thermal stress in larvae of the reef-building coral Montastraea faveolata. PLoS ONE 2010, 5, e11221. [Google Scholar] [CrossRef] [PubMed]
- Gallagher, P.J.; Herring, B.P.; Stull, J.T. Myosin light chain kinase. J. Muscle Res. Cell Motil. 1997, 18, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Farah, M.E.; Sirotkin, V.; Haarer, B.; Kakhniashvili, D.; Amberg, D.C. Diverse protective roles of the actin cytoskeleton during oxidative stress. Cytoskeleton 2011, 68, 340–354. [Google Scholar] [CrossRef] [PubMed]
- Larsen, M.; Artym, V.V.; Green, J.A.; Yamada, K.M. The matrix reorganized: Extracellular matrix remodeling and integrin signaling. Curr. Opin. Cell Biol. 2006, 18, 463–471. [Google Scholar] [CrossRef] [PubMed]
- NCBI Sequence Read Archive. Available online: https://trace.ncbi.nlm.nih.gov/Traces/sra/sra.cgi? (accessed on 1 January 2018).
- Gene Ontology Database. Available online: http://www.geneontology.org/ (accessed on 8 January 2018).
- Kyoto Encyclopedia of Genes and Genomes Database. Available online: http://www.genome.jp/kegg/ (accessed on 8 January 2018).
Sample Availability: Samples of the compounds are not available from the authors. |




| Sample | Raw Data | Clean Data | Valid Ratio % | Q20 % | Q30 % | GC Content % | ||
|---|---|---|---|---|---|---|---|---|
| Read | Base | Read | Base | Read | ||||
| CON_1 | 53397180 | 8.01G | 52327150 | 7.85G | 98.00 | 99.40 | 93.73 | 42 |
| CON_2 | 54956050 | 8.24G | 53785462 | 8.07G | 97.87 | 99.57 | 94.67 | 41 |
| CON_3 | 58609160 | 8.79G | 57471652 | 8.62G | 98.06 | 99.25 | 92.29 | 40 |
| T_1h_1 | 48381506 | 7.26G | 47514898 | 7.13G | 98.21 | 99.57 | 94.62 | 43 |
| T_1h_2 | 56821096 | 8.52G | 55435722 | 8.32G | 97.56 | 99.02 | 91.34 | 41 |
| T_1h_3 | 55081090 | 8.26G | 53965518 | 8.09G | 97.97 | 99.38 | 93.39 | 42 |
| T_3h_1 | 50974702 | 7.65G | 50010072 | 7.50G | 98.11 | 99.20 | 92.92 | 41 |
| T_3h_2 | 56378238 | 8.46G | 54928996 | 8.24G | 97.43 | 99.42 | 94.27 | 42 |
| T_3h_3 | 51550962 | 7.73G | 50534502 | 7.58G | 98.03 | 99.38 | 92.95 | 43 |
| Sample | Valid Reads | Mapped Reads | Unique Mapped Reads | Multimapped Reads |
|---|---|---|---|---|
| CON_1 | 52327150 | 40400777(77.21%) | 22317749(42.65%) | 18083028(34.56%) |
| CON_2 | 53785462 | 40377886(75.07%) | 23759345(44.17%) | 16618541(30.90%) |
| CON_3 | 57471652 | 41429455(72.09%) | 24918806(43.36%) | 16510649(28.73%) |
| T_1h_1 | 47514898 | 33468336(70.44%) | 18785041(39.54%) | 14683295(30.90%) |
| T_1h_2 | 55435722 | 41648386(75.13%) | 23013370(41.51%) | 18635016(33.62%) |
| T_1h_3 | 53965518 | 40764443(75.54%) | 24024127(44.52%) | 16740316(31.02%) |
| T_3h_1 | 50010072 | 38674656(77.33%) | 16814094(33.62%) | 21860562(43.71%) |
| T_3h_2 | 54928996 | 45056381(82.03%) | 22811470(41.53%) | 22244911(40.50%) |
| T_3h_3 | 50534502 | 36913545(73.05%) | 22295211(44.12%) | 14618334(28.93%) |
| Gene ID | Gene Name | T_1h vs. CON | T_3h vs. CON | ||
|---|---|---|---|---|---|
| log2(fc) | p | log2 (fc) | p | ||
| Endogenous Protease | |||||
| MSTRG.28936 | 72 kDa type IV collagenase | 1.92 | 0.03 | 2.22 | 0.03 |
| MSTRG.5390 | MMP 16 precursor | 1.57 | 0.04 | 1.50 | 0.03 |
| Oxidative Stress Response | |||||
| MSTRG.45929 | Glutathione S-transferase theta-1-like | 1.20 | 0.04 | 1.68 | 0.01 |
| MSTRG.38457 | Microsomal glutathione S-transferase 1 | −0.35 | 0.18 | −0.99 | 0.02 |
| MSTRG.1616 | Glutathione peroxidase | 1.08 | 0.02 | 1.75 | 0.02 |
| Immune System Process | |||||
| MSTRG.9314 | Fibrinogen-like protein A | 2.00 | 0.00 | 1.57 | 0.04 |
| MSTRG.14673 | Fibrinogen C domain-containing protein 1-like | 0.76 | 0.29 | 1.04 | 0.03 |
| MSTRG.30809 | Ficolin-2 | −1.51 | 0.29 | −2.49 | 0.03 |
| MSTRG.40130 | Tenascin-N | −1.93 | 0.05 | 0.93 | 0.63 |
| MSTRG.13644 | Complement component C3 | 3.29 | 0.02 | 3.19 | 0.01 |
| MSTRG.2787 | Complement factor B | 2.49 | 0.00 | 2.24 | 0.03 |
| MSTRG.3468 | Complement factor B-2 | 1.81 | 0.17 | 2.02 | 0.04 |
| MSTRG.16733 | Mannan-binding lectin serine protease 1-like | 1.39 | 0.00 | 1.45 | 0.03 |
| MSTRG.25958 | Lactose-binding lectin l-2-like | 1.58 | 0.03 | 1.21 | 0.16 |
| MSTRG.37300 | HSP70 | 1.14 | 0.04 | 1.87 | 0.00 |
| MSTRG.6868 | Integrin beta-like protein A | 3.63 | 0.00 | 3.47 | 0.00 |
| MSTRG.21840 | Integrin alpha-4 | −0.69 | 0.14 | −1.50 | 0.05 |
| Apoptosis | |||||
| MSTRG.20169 | PUF60 isoform X3 | 1.20 | 0.04 | 1.04 | 0.19 |
| MSTRG.4067 | TFIIH basal transcription factor complex helicase XPB subunit-like | 0.67 | 0.09 | 1.33 | 0.02 |
| MSTRG.14930 | Bax inhibitor-1 | −0.65 | 0.02 | −1.30 | 0.03 |
| MSTRG.9235 | Src family kinase 5 | −0.72 | 0.05 | −1.14 | 0.04 |
| Cytoskeleton and ECM associated genes | |||||
| MSTRG.2582 | Myosin heavy chain, striated muscle isoform X5 | 1.72 | 0.03 | 2.14 | 0.00 |
| MSTRG.23618 | Myosin heavy chain, striated muscle isoform X7 | 2.25 | 0.01 | 2.39 | 0.01 |
| MSTRG.21690 | Myosin-10 | 1.07 | 0.01 | 0.61 | 0.00 |
| MSTRG.13638 | Unconventional myosin-X | 0.65 | 0.11 | 1.16 | 0.03 |
| MSTRG.36459 | Myosin-2 essential light chain-like | 1.34 | 0.15 | 2.33 | 0.01 |
| MSTRG.2896 | Myosin light chain kinase, smooth muscle | 2.03 | 0.06 | 2.35 | 0.01 |
| MSTRG.23243 | Actin isoform 2 | 1.34 | 0.11 | 1.65 | 0.01 |
| MSTRG.7111 | Actin | −0.40 | 0.55 | −1.14 | 0.00 |
| MSTRG.30094 | Titin | 2.10 | 0.02 | 3.00 | 0.01 |
| MSTRG.18253 | Troponin I, partial | 1.64 | 0.03 | 1.93 | 0.01 |
| MSTRG.29309 | Tropomodulin-1-like | −0.57 | 0.13 | −1.10 | 0.04 |
| MSTRG.30449 | Gelsolin-like protein 2 | 1.23 | 0.17 | 2.04 | 0.00 |
| MSTRG.37982 | Alpha-1 collagen isoform X4 | 1.27 | 0.04 | 1.63 | 0.04 |
| MSTRG.5905 | Alpha-2 collagen | 1.75 | 0.03 | 2.48 | 0.01 |
| MSTRG.28962 | Collagen alpha-1(V) chain | 1.33 | 0.00 | 1.29 | 0.15 |
| MSTRG.41401 | Collagen IV alpha-3-binding protein-like | 1.38 | 0.04 | 1.22 | 0.21 |
| Gene ID | T_1h vs. CON | T_3h vs. CON | ||
|---|---|---|---|---|
| RNA-Seq | RT-qPCR | RNA-Seq | RT-qPCR | |
| MSTRG.6868 | 3.63 ** | 1.75 ** | 3.47 ** | 2.69 ** |
| MSTRG.13644 | 3.29 * | 2.95 ** | 3.19 * | 2.03 ** |
| MSTRG.23313 | 1.97 ** | 2.26 * | 2.04 * | 5.55 ** |
| MSTRG.32095 | 1.94 ** | 3.05 ** | 1.77 ** | 3.51 * |
| MSTRG.28936 | 1.92 * | 1.55 * | 2.22 * | 1.26 * |
| MSTRG.22626 | 1.84 * | 2.15 * | 1.87 * | 3.47 ** |
| MSTRG.18184 | 1.67 ** | 2.14 ** | 1.38 * | 4.71 ** |
| MSTRG.5390 | 1.57 * | 1.23 * | 1.50 * | 2.01 * |
| MSTRG.16733 | 1.39 ** | 1.67 * | 1.45 * | 2.31 ** |
| MSTRG.37982 | 1.27 * | 2.00 * | 1.63 * | 9.68 ** |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dong, X.; Qi, H.; He, B.; Jiang, D.; Zhu, B. RNA Sequencing Analysis to Capture the Transcriptome Landscape during Tenderization in Sea Cucumber Apostichopus japonicus. Molecules 2019, 24, 998. https://doi.org/10.3390/molecules24050998
Dong X, Qi H, He B, Jiang D, Zhu B. RNA Sequencing Analysis to Capture the Transcriptome Landscape during Tenderization in Sea Cucumber Apostichopus japonicus. Molecules. 2019; 24(5):998. https://doi.org/10.3390/molecules24050998
Chicago/Turabian StyleDong, Xiufang, Hang Qi, Baoyu He, Di Jiang, and Beiwei Zhu. 2019. "RNA Sequencing Analysis to Capture the Transcriptome Landscape during Tenderization in Sea Cucumber Apostichopus japonicus" Molecules 24, no. 5: 998. https://doi.org/10.3390/molecules24050998
APA StyleDong, X., Qi, H., He, B., Jiang, D., & Zhu, B. (2019). RNA Sequencing Analysis to Capture the Transcriptome Landscape during Tenderization in Sea Cucumber Apostichopus japonicus. Molecules, 24(5), 998. https://doi.org/10.3390/molecules24050998