The Effect of Rhodamine-Derived Superparamagnetic Maghemite Nanoparticles on the Motility of Human Mesenchymal Stem Cells and Mouse Embryonic Fibroblast Cells
Abstract
:1. Introduction
2. Results
2.1. Cell Morphology and Flow Cytometry
2.2. Quantification of Reactive Oxygen Species (ROS) Generation after SAMN-R Labelling
2.3. Cell Proliferation
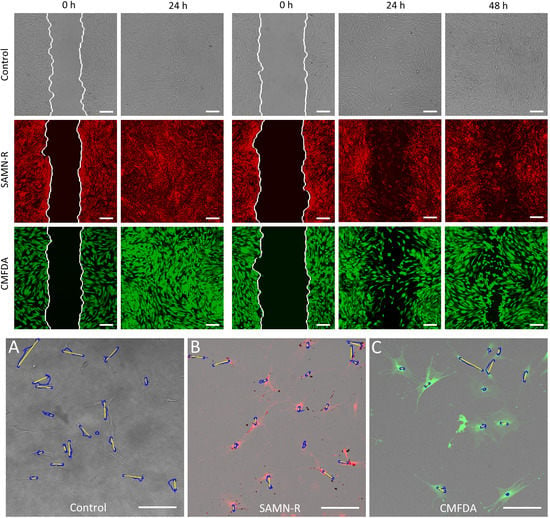
2.4. Cell Migration Study
2.4.1. In Vitro Wound Healing Assay
2.4.2. Single-Cell Migration
3. Discussion
4. Materials and Methods
4.1. Synthesis and Characterisation of SAMN-R
4.2. Cell Cultures
4.3. Cell Labelling
4.4. Confocal Microscopy
4.5. Cell Morphology and Flow Cytometry
4.6. Quantification of ROS Generation after SAMN-R Labelling
4.7. Cell Growth Curves
4.8. Analysis of Wound Healing Assay
4.9. Analysis of Single-Cell Migration
4.10. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Ramaswamy, S.; Greco, J.B.; Uluer, M.C.; Zhang, Z.; Zhang, Z.; Fishbein, K.W.; Spencer, R.G. Magnetic Resonance Imaging of Chondrocytes Labeled with Superparamagnetic Iron Oxide Nanoparticles in Tissue-Engineered Cartilage. Tissue Eng. Part A 2009, 15, 3899–3910. [Google Scholar] [CrossRef] [Green Version]
- Sun, J.; Zhou, S.; Hou, P.; Yang, Y.; Weng, J.; Li, X.; Li, M. Synthesis and Characterization of Biocompatible Fe3O4 Nanoparticles. J. Biomed. Mater. Res. Part A 2007, 80A, 333–341. [Google Scholar] [CrossRef]
- Kyrtatos, P.G.; Lehtolainen, P.; Junemann-Ramirez, M.; Garcia-Prieto, A.; Price, A.N.; Martin, J.F.; Gadian, D.G.; Pankhurst, Q.A.; Lythgoe, M.F. Magnetic Tagging increases Delivery of Circulating Progenitors in Vascular injury. JACC: Cardiovasc. Interv. 2009, 2, 794–802. [Google Scholar] [CrossRef] [PubMed]
- Ito, A.; Hibino, E.; Kobayashi, C.; Terasaki, H.; Kagami, H.; Ueda, M.; Kobayashi, T.; Honda, H. Construction and Delivery of Tissue-Engineered Human Retinal Pigment Epithelial Cell Sheets, Using Magnetite Nanoparticles and Magnetic Force. Tissue Eng. 2005, 11, 489–496. [Google Scholar] [CrossRef] [PubMed]
- Scherer, F.; Anton, M.; Schillinger, U.; Henke, J.; Bergemann, C.; Krüger, A.; Gänsbacher, B.; Plank, C. Magnetofection: Enhancing and Targeting Gene Delivery by Magnetic Force in vitro and in vivo. Gene Ther. 2002, 9, 102–109. [Google Scholar] [CrossRef] [PubMed]
- Reddy, L.H.; Arias, J.L.; Nicolas, J.; Couvreur, P. Magnetic Nanoparticles: Design and Characterization, Toxicity and Biocompatibility, Pharmaceutical and Biomedical Applications. Chem. Rev. 2012, 112, 5818–5878. [Google Scholar] [CrossRef] [PubMed]
- Cmiel, V.; Skopalik, J.; Polakova, K.; Solar, J.; Havrdova, M.; Milde, D.; Justan, I.; Magro, M.; Starcuk, Z.; Provaznik, I. Rhodamine Bound Maghemite As A Long-Term Dual Imaging Nanoprobe of Adipose Tissue-Derived Mesenchymal Stromal Cells. Eur. Biophys. J. 2017, 46, 433–444. [Google Scholar] [CrossRef]
- Skopalik, J.; Polakova, K.; Havrdova, M.; Justan, I.; Magro, M.; Milde, D.; Knopfova, L.; Smarda, J.; Polakova, H.; Gabrielova, E.; et al. Mesenchymal Stromal Cell Labeling by New Uncoated Superparamagnetic Maghemite Nanoparticles in Comparison with Commercial Resovist—An initial in vitro Study. Int. J. Nanomed. 2014, 2014, 5355–5372. [Google Scholar] [CrossRef]
- Kong, B.; Seog, J.H.; Graham, L.M.; Lee, S.B. Experimental Considerations on the Cytotoxicity of Nanoparticles. Nanomedicine 2011, 6, 929–941. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Liu, J.; Zheng, H.; Wichmann, J.; Hopfner, U.; Sudhop, S.; Prein, C.; Shen, Y.; Machens, H.-G.; Schilling, A.F. Nano-Formulated Curcumin Accelerates Acute Wound Healing Through Dkk-1-Mediated Fibroblast Mobilization and Mcp-1-Mediated Anti-inflammation. NPG Asia Mater. 2017, 9, e368. [Google Scholar] [CrossRef]
- Haubner, F.; Muschter, D.; Pohl, F.; Schreml, S.; Prantl, L.; Gassner, H. A Co-Culture Model of Fibroblasts and Adipose Tissue-Derived Stem Cells Reveals New insights into Impaired Wound Healing After Radiotherapy. Int. J. Mol. Sci. 2015, 16, 25947–25958. [Google Scholar] [CrossRef] [PubMed]
- Magro, M.; Sinigaglia, G.; Nodari, L.; Tucek, J.; Polakova, K.; Marusak, Z.; Cardillo, S.; Salviulo, G.; Russo, U.; Stevanato, R.; et al. Charge Binding of Rhodamine Derivative TO Oh−Stabilized Nanomaghemite: Universal Nanocarrier For Construction of Magnetofluorescent Biosensors. Acta Biomater. 2012, 8, 2068–2076. [Google Scholar] [CrossRef] [PubMed]
- Ettinger, A.; Wittmann, T. Fluorescence Live Cell Imaging. In Quantitative Imaging in Cell Biology; Methods in Cell Biology; Elsevier: Amsterdam, The Netherlands, 2014; pp. 77–94. [Google Scholar]
- Jensen, E.C. Use of Fluorescent Probes: Their Effect on Cell Biology and Limitations. Anat. Record 2012, 295, 2031–2036. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kramer, N.; Walzl, A.; Unger, C.; Rosner, M.; Krupitza, G.; Hengstschläger, M.; Dolznig, H. in vitro Cell Migration and invasion Assays. Mutat. Res./Rev. Mutat. Res. 2013, 752, 10–24. [Google Scholar] [CrossRef] [PubMed]
- Fronza, M.; Heinzmann, B.; Hamburger, M.; Laufer, S.; Merfort, I. Determination of the Wound Healing Effect of Calendula Extracts Using the Scratch Assay With 3T3 Fibroblasts. J. Ethnopharmacol. 2009, 126, 463–467. [Google Scholar] [CrossRef] [PubMed]
- Yeom, C.-H.; Lee, G.; Park, J.-H.; Yu, J.; Park, S.; Yi, S.-Y.; Lee, H.; Hong, Y.; Yang, J.; Lee, S. High Dose Concentration Administration of Ascorbic Acid inhibits Tumor Growth in Balb/c Mice Implanted with Sarcoma 180 Cancer Cells Via the Restriction of Angiogenesis. J. Transl. Med. 2009, 7, 1–9. [Google Scholar] [CrossRef]
- Liu, Q.; Xu, Y.; Wei, S.; Gao, W.; Chen, L.; Zhou, T.; Wang, Z.; Ying, M.; Zheng, Q. Mirna-148B Suppresses Hepatic Cancer Stem Cell by Targeting Neuropilin-1. Biosci. Rep. 2015, 35, e00229. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.-C.; Park, A.Y.; Guan, J.-L. in vitro Scratch Assay: A Convenient and inexpensive Method for Analysis of Cell Migration in vitro. Nat. Protoc. 2007, 2, 329–333. [Google Scholar] [CrossRef]
- De Pascalis, C.; Etienne-Manneville, S.; Weaver, V.M. Single and Collective Cell Migration: the Mechanics of Adhesions. Mol. Biol. Cell. 2017, 28, 1833–1846. [Google Scholar] [CrossRef] [PubMed]
- Lintz, M.; Muñoz, A.; Reinhart-King, C.A. the Mechanics of Single Cell and Collective Migration of Tumor Cells. J. Biomech. Eng. 2017, 139, 1–9. [Google Scholar] [CrossRef]
- Jin, C.-Y.; Zhu, B.-S.; Wang, X.-F.; Lu, Q.-H. Cytotoxicity of Titanium Dioxide Nanoparticles in Mouse Fibroblast Cells. Chem. Res. Toxicol. 2008, 21, 1871–1877. [Google Scholar] [CrossRef] [PubMed]
- Coradeghini, R.; Gioria, S.; García, C.P.; Nativo, P.; Franchini, F.; Gilliland, D.; Ponti, J.; Rossi, F. Size-Dependent Toxicity and Cell interaction Mechanisms of Gold Nanoparticles on Mouse Fibroblasts. Toxicol. Lett. 2013, 217, 205–216. [Google Scholar] [CrossRef]
- Vikesland, P.J.; Rebodos, R.L.; Bottero, J.Y.; Rose, J.; Masion, A. Aggregation and Sedimentation of Magnetite Nanoparticle Clusters. Environ. Sci. Nano 2016, 3, 567–577. [Google Scholar] [CrossRef]
- Pernodet, N.; Fang, X.; Sun, Y.; Bakhtina, A.; Ramakrishnan, A.; Sokolov, J.; Ulman, A.; Rafailovich, M. Adverse Effects of Citrate/gold Nanoparticles on Human Dermal Fibroblasts. Small 2006, 2, 766–773. [Google Scholar] [CrossRef] [PubMed]
- Berry, C.C.; Wells, S.; Charles, S.; Aitchison, G.; Curtis, A.S.G. Cell Response to Dextran-Derivatised Iron Oxide Nanoparticles Post internalisation. Biomaterials 2004, 25, 5405–5413. [Google Scholar] [CrossRef]
- Wu, X.; Tan, Y.; Mao, H.; Zhang, M. Toxic Effects of Iron Oxide Nanoparticles on Human Umbilical Vein Endothelial Cells. Int. J. Nanomed. 2010, 2010, 385–399. [Google Scholar] [CrossRef]
- Cromer Berman, S.M.; Kshitiz; Wang, C.J.; Orukari, I.; Levchenko, A.; Bulte, J.W.M.; Walczak, P. Cell Motility of Neural Stem Cells Is Reduced After Spio-Labeling, Which Is Mitigated After Exocytosis. Magn. Reson. Med. 2013, 69, 255–262. [Google Scholar] [CrossRef]
- Tay, C.Y.; Cai, P.; Setyawati, M.I.; Fang, W.; Tan, L.P.; Hong, C.H.L.; Chen, X.; Leong, D.T. Nanoparticles Strengthen intracellular Tension and Retard Cellular Migration. Nano Lett. 2013, 14, 83–88. [Google Scholar] [CrossRef]
- Yañez, R.; Lamana, M.L.; García-Castro, J.; Colmenero, I.; Ramírez, M.; Bueren, J.A. Adipose Tissue-Derived Mesenchymal Stem Cells Have in vivo Immunosuppressive Properties Applicable For the Control of the Graft-Versus-Host Disease. Stem Cells 2006, 24, 2582–2591. [Google Scholar] [CrossRef]
- Baiazitova, L.; Skopalik, J.; Cmiel, V.; Chmelik, J.; Svoboda, O.; Provaznik, I. Modern Semi-Automatic Set-Up for Testing Cell Migration with Impact for Therapy of Myocardial infarction. In World Congress on Medical Physics and Biomedical Engineering 2018; IFMBE Proceedings; Springer: Singapore, 2019; pp. 155–159. [Google Scholar]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An Open-Source Platform for Biological-Image Analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- Rueden, C.T.; Schindelin, J.; Hiner, M.C.; DeZonia, B.E.; Walter, A.E.; Arena, E.T.; Eliceiri, K.W. Imagej2: Imagej For the Next Generation of Scientific Image Data. BMC Bioinform. 2017, 18, 529. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the SAMN-R are available from the authors. |











| Item | Time | Control | SAMN-R | CMFDA | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Min | Max | Mean | Min | Max | Mean | Min | Max | Mean | ||
| Scratch width (µm) | 311.54 | 493.62 | 408.01 | 325.04 | 477.73 | 416.44 | 329.51 | 491.50 | 427.76 | |
| Scratch area (mm2) | 0 h | 0.36 | 0.58 | 0.48 | 0.38 | 0.56 | 0.49 | 0.38 | 0.57 | 0.50 |
| 24 h | 0.00 | 0.02 | 0.00 | 0.00 | 0.02 | 0.00 | 0.00 | 0.02 | 0.00 | |
| Item | Time | Control | SAMN-R | CMFDA | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Min | Max | Mean | Min | Max | Mean | Min | Max | Mean | ||
| Scratch width (µm) | 502.18 | 689.57 | 554.24 | 438.18 | 711.15 | 548.73 | 447.23 | 621.20 | 541.37 | |
| Scratch area (mm2) | 0 h | 0.59 | 0.80 | 0.65 | 0.51 | 0.83 | 0.64 | 0.52 | 0.73 | 0.63 |
| 24 h | 0.17 | 0.53 | 0.31 | 0.14 | 0.57 | 0.32 | 0.16 | 0.56 | 0.36 | |
| 48 h | 0.00 | 0.09 | 0.03 | 0.00 | 0.05 | 0.02 | 0.00 | 0.23 | 0.03 | |
| Control | SAMN-R | CMFDA | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Min | Max | Mean | Min | Max | Mean | Min | Max | Mean | |
| Accumulated distance (µm) | 62.76 | 271.85 | 153.99 | 86.36 | 241.65 | 148.85 | 73.92 | 200.72 | 136.70 |
| Euclidean distance (µm) | 1..23 | 217.19 | 81.90 | 8.83 | 123.17 | 55.36 | 7.91 | 97.74 | 41.92 |
| Velocity (µm/min) | 0.17 | 0.76 | 0.43 | 0.24 | 0.67 | 0.41 | 0.21 | 0.56 | 0.38 |
| Control | SAMN-R | CMFDA | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Min | Max | Mean | Min | Max | Mean | Min | Max | Mean | |
| Accumulated distance (µm) | 32.82 | 294.80 | 104.90 | 44.64 | 362.05 | 118.83 | 51.73 | 221.71 | 114.01 |
| Euclidean distance (µm) | 7.24 | 248.21 | 69.32 | 6.87 | 316.28 | 80.86 | 12.22 | 194.51 | 82.98 |
| Velocity (µm/min) | 0.09 | 0.82 | 0.29 | 0.12 | 1.01 | 0.33 | 0.14 | 0.62 | 0.32 |
| Control | SAMN-R | CMFDA | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Min | Max | Mean | Min | Max | Mean | Min | Max | Mean | |
| Accumulated distance (µm) | 48.06 | 288.48 | 156.00 | 45.15 | 270.70 | 143.62 | 43.02 | 282.62 | 154.66 |
| Euclidean distance (µm) | 9.72 | 190.26 | 65.91 | 15.74 | 226.10 | 99.33 | 2.26 | 193.26 | 68.11 |
| Velocity (µm/min) | 0.13 | 0.80 | 0.43 | 0.13 | 0.75 | 0.40 | 0.12 | 0.79 | 0.43 |
| Control | SAMN-R | CMFDA | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Min | Max | Mean | Min | Max | Mean | Min | Max | Mean | |
| Accumulated distance (µm) | 33.29 | 179.74 | 90.96 | 30.09 | 181.34 | 84.78 | 37.08 | 263.26 | 79.96 |
| Euclidean distance (µm) | 9.71 | 147.88 | 61.31 | 8.03 | 102.06 | 40.66 | 3.41 | 243.35 | 42.51 |
| Velocity (µm/min) | 0.09 | 0.50 | 0.25 | 0.08 | 0.50 | 0.24 | 0.10 | 0.73 | 0.22 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baiazitova, L.; Skopalik, J.; Chmelik, J.; Zumberg, I.; Cmiel, V.; Polakova, K.; Provaznik, I. The Effect of Rhodamine-Derived Superparamagnetic Maghemite Nanoparticles on the Motility of Human Mesenchymal Stem Cells and Mouse Embryonic Fibroblast Cells. Molecules 2019, 24, 1192. https://doi.org/10.3390/molecules24071192
Baiazitova L, Skopalik J, Chmelik J, Zumberg I, Cmiel V, Polakova K, Provaznik I. The Effect of Rhodamine-Derived Superparamagnetic Maghemite Nanoparticles on the Motility of Human Mesenchymal Stem Cells and Mouse Embryonic Fibroblast Cells. Molecules. 2019; 24(7):1192. https://doi.org/10.3390/molecules24071192
Chicago/Turabian StyleBaiazitova, Larisa, Josef Skopalik, Jiri Chmelik, Inna Zumberg, Vratislav Cmiel, Katerina Polakova, and Ivo Provaznik. 2019. "The Effect of Rhodamine-Derived Superparamagnetic Maghemite Nanoparticles on the Motility of Human Mesenchymal Stem Cells and Mouse Embryonic Fibroblast Cells" Molecules 24, no. 7: 1192. https://doi.org/10.3390/molecules24071192
APA StyleBaiazitova, L., Skopalik, J., Chmelik, J., Zumberg, I., Cmiel, V., Polakova, K., & Provaznik, I. (2019). The Effect of Rhodamine-Derived Superparamagnetic Maghemite Nanoparticles on the Motility of Human Mesenchymal Stem Cells and Mouse Embryonic Fibroblast Cells. Molecules, 24(7), 1192. https://doi.org/10.3390/molecules24071192