Coralmycin Derivatives with Potent Anti-Gram Negative Activity Produced by the Myxobacteria Corallococcus coralloides M23
Abstract
:1. Introduction
2. Results and Discussion
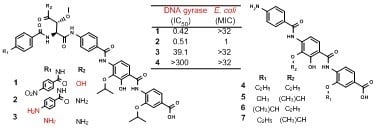
2.1. Structural Elucidation
2.2. Antibacterial and E. Coli DNA Gyrase-Inhibitory Activities of the New Compounds
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Fermentation and Isolation
3.3. Determination of Antibacterial Susceptibility
3.4. DNA Gyrase Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Boucher, H.W.; Talbot, G.H.; Benjamin, D.K., Jr.; Bradley, J.; Guidos, R.J.; Jones, R.N.; Murray, B.E.; Bonomo, R.A.; Gilbert, D. 10 × ’20 Progress–development of new drugs active against gram-negative bacilli: An update from the Infectious Diseases Society of America. Clin. Infect. Dis. 2013, 56, 1685–1694. [Google Scholar] [CrossRef] [PubMed]
- Giske, C.G.; Monnet, D.L.; Cars, O.; Carmeli, Y. Clinical and economic impact of common multidrug-resistant gram-negative bacilli. Antimicrob. Agents Chemother. 2008, 52, 813–821. [Google Scholar] [CrossRef] [PubMed]
- Boucher, H.W.; Talbot, G.H.; Bradley, J.S.; Edwards, J.E.; Gilbert, D.; Rice, L.B.; Scheld, M.; Spellberg, B.; Bartlett, J. Bad bugs, no drugs: No ESKAPE! An update from the Infectious Diseases Society of America. Clin. Infect. Dis. 2009, 48, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.J.; Kim, H.J.; Kim, G.W.; Cho, K.; Takahashi, S.; Koshino, H.; Kim, W.G. Isolation of Coralmycins A and B, Potent Anti-Gram Negative Compounds from the Myxobacteria Corallococcus coralloides M23. J. Nat. Prod. 2016, 79, 2223–2228. [Google Scholar] [CrossRef] [PubMed]
- Baumann, S.; Herrmann, J.; Raju, R.; Steinmetz, H.; Mohr, K.I.; Huttel, S.; Harmrolfs, K.; Stadler, M.; Muller, R. Cystobactamids: Myxobacterial topoisomerase inhibitors exhibiting potent antibacterial activity. Angew. Chem. Int. Ed. Engl. 2014, 53, 14605–14609. [Google Scholar] [CrossRef] [PubMed]
- Cociancich, S.; Pesic, A.; Petras, D.; Uhlmann, S.; Kretz, J.; Schubert, V.; Vieweg, L.; Duplan, S.; Marguerettaz, M.; Noell, J.; et al. The gyrase inhibitor albicidin consists of p-aminobenzoic acids and cyanoalanine. Nat. Chem. Biol. 2015, 11, 195–197. [Google Scholar] [CrossRef] [PubMed]
- Huttel, S.; Testolin, G.; Herrmann, J.; Planke, T.; Gille, F.; Moreno, M.; Stadler, M.; Bronstrup, M.; Kirschning, A.; Muller, R. Discovery and Total Synthesis of Natural Cystobactamid Derivatives with Superior Activity against Gram-Negative Pathogens. Angew. Chem. Int. Ed. Engl. 2017, 56, 12760–12764. [Google Scholar] [CrossRef] [PubMed]
- Pouchert, C.J.; Behnke, J. The Aldrich Library of ¹³C and ¹H FT NMR Spectra; Aldrich Chemical Co.: Milwaukee, WI, USA, 1993; p. 1085. [Google Scholar]
- Cheng, B.; Muller, R.; Trauner, D. Total Syntheses of Cystobactamids and Structural Confirmation of Cystobactamid 919-2. Angew. Chem. Int. Ed. Engl. 2017, 56, 12755–12759. [Google Scholar] [CrossRef] [PubMed]
- Zheng, C.J.; Sohn, M.J.; Lee, S.; Kim, W.G. Meleagrin, a new FabI inhibitor from Penicillium chryosogenum with at least one additional mode of action. PLoS ONE 2013, 8, e78922. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compound 11 are available from the authors. |



| Position | 11 | 1 | 2 | 3 | 8 | 9 |
|---|---|---|---|---|---|---|
| 1 | - | - | - | - | - | - |
| 2 | - | - | - | - | - | - |
| 3 | 7.56, brs | 7.57, s | 7.59, s | 7.58 * | 7.57, s | 7.55, s |
| 4 | - | - | - | - | - | - |
| 5 | - | - | - | - | - | - |
| 6 | 8.50, d (8.3) | 8.51, d (8.3) | 8.52, d (8.4) | 8.51, d (8.2) | 8.39, d (8.3) | 8.46, d (8.3) |
| 7 | 7.58, d (8.3) | 7.59, d (8.7) | 7.60, d (8.3) | 7.58 * | 7.62, d (8.3) | 7.61, d (8.3) |
| 8 | 4.75, m | 4.75, m | 4.76, m | 4.75, m | 3.97, s | 4.2, q (7.0) |
| 9,10 | 1.37, d (6.0) | 1.37, d (6.0) | 1.38, d (6.0) | 1.37, d (5.8) | - | 1.46, t (7.1) |
| 11-NH | 10.97, brs | - | 11.00, s | 11.00, s | 10.89, s | 10.96, s |
| 1′ | - | - | - | - | - | - |
| 2′ | - | - | - | - | - | - |
| 3′ | - | - | - | - | - | - |
| 3′-OH | 11.21, brs | 11.30, s | 11.27, s | 11.27, s | 11.46, s | 11.31, s |
| 4′ | - | - | - | - | - | - |
| 5′ | - | - | - | - | - | - |
| 6′ | 7.50, d (8.5) | 7.52, d (8.7) | 7.54, d (8.8) | 7.52, d (8.7) | 7.55, m | 7.54, d (8.7) |
| 7′ | 7.80, d (8.5) | 7.81, d (8.7) | 7.84 * | 7.81 * | 7.82, m | 7.82, d (7.6) |
| 8′ | 4.32, m | 4.31, m | 4.33, m | 4.30, m | 4.38, m | 4.33, m |
| 9′,10′ | 1.26, d (6.1) | 1.26, d (6.1) | 1.28, d (6.1) | 1.26, d (6.0) | 1.27, d (6.4) | 1.26, d (6.1) |
| 11′-NH | 9.58, s | 9.60, s | 9.61, s | 9.60, s | 9.58, s | 9.59, s |
| 1′′ | - | - | - | - | - | - |
| 2′′ | - | - | - | - | - | - |
| 3′′,7′′ | 7.97, d (8.6) | 7.97, d (8.5) | 7.98, d (8.8) | 7.96, (8.3) | 7.98, d (8.7) | 7.97, d (8.8) |
| 4′′,6′′ | 7.83, d (8.6) | 7.84, d (8.5) | 7.84 * | 7.81 * | 7.83, m | 7.83, d (7.6) |
| 5′′ | - | - | - | - | - | - |
| 8′′-NH | 10.56, s | 10.52, s | 10.57, s | 10.50, s | 10.57, s | 10.57, s |
| 1′′′ | - | - | - | - | - | - |
| 2′′′ | 4.92, dd (8.0, 8.1) | 5.07, t (8.3) | 4.92, t (8.1) | 4.82, t (7.9) | 4.92, t (8.1) | 4.92, t (8.1) |
| 3′′′ | 4.09, d (8.0) | 4.16, d (8.1) | 4.11, d (8.0) | 4.06, d (8.0) | 4.09, d (8.1) | 4.09, d (8.1) |
| 4′′′-NH | 7.47, brs 7.54, brs | - | 7.48, brs | 7.45, brs | 7.48, brs | 7.48, brs |
| 5′′′ | 3.31, s | 3.35, s | 3.32, s | 3.28, s | 3.31, s | 3.31, s |
| 6′′′-NH | 8.46, d (8.1) | 8.72, d (8.5) | 8.41, d (8.2) | 8.00, d (8.0) | 8.47, d (8.1) | 8.47, d (8.1) |
| 1′′′′ | - | - | - | - | - | - |
| 2′′′′ | - | - | - | - | - | - |
| 3′′′′,7′′′′ | 7.90, d * | 7.94, d (8.6) | 7.84 * | 7.58 * | 7.91 * | 7.91 * |
| 4′′′′,6′′′′ | 7.90, d * | 7.90, d (8.6) | 7.84 * | 6.57, d (8.1) | 7.91* | 7.91* |
| 5′′′′ | - | - | - | - | - | - |
| 8′′′′-NH | 10.8, s | 10.84, s | 10.02, s | - | 10.81, s | 10.81, s |
| 1′′′′′ | - | - | - | - | - | - |
| 2′′′′′ | - | - | - | - | - | - |
| 3′′′′′,7′′′′′ | 8.21, d (8.6) | 8.21, d (8.6) | 7.75, d (8.8) | - | 8.21, d (8.8) | 8.21, d (8.8) |
| 4′′′′′,6′′′′′ | 8.38, d (8.6) | 8.38, d (8.6) | 6.62, d (8.6) | - | 8.39, d (8.8) | 8.39, d (8.8) |
| 5′′′′′ | - | - | - | - | - | - |
| Position | 11 | 1 | 2 | 3 | 8 | 9 |
|---|---|---|---|---|---|---|
| 1 | 166.9, C | 167.4, C | 167.3, C | 167.0, C | 166.8, C | 166.9, C |
| 2 | 125.7, C | 126.2, C | 126.2, C | 125.8, C | 126.1, C | 126.3, C |
| 3 | 113.9, CH | 114.3, CH | 114.4, CH | 113.9, CH | 111.7, CH | 111.9, CH |
| 4 | 146.3, C | 146.8, C | 146.8, C | 146.4, C | 148.4, C | 148.0, C |
| 5 | 133.3, C | 133.7, C | 133.4, C | 133.3, C | 131.4, C | 132.6, C |
| 6 | 119.6, CH | 120.0, CH | 120.1, CH | 119.6, CH | 120.4, CH | 119.6, CH |
| 7 | 122.6, CH | 123.1, CH | 123.0, CH | 122.7, CH | 122.4, CH | 122.6, CH |
| 8 | 71.7, CH | 72.1, CH | 72.2, CH | 71.8, CH | 56.5, CH3 | 64.1, CH2 |
| 9,10 | 21.6, CH3 | 22.0, CH3 | 22.0, CH3 | 21.7, CH3 | - | 14.3, CH3 |
| 1′ | 163.6, C | 164.0, C | 164.0, C | 163.6, C | 164.2, C | 136.6, C |
| 2′ | 116.4, C | 116.9, C | 117.0, C | 116.6, C | 115.5, C | 116.4, C |
| 3′ | 150.3, C | 150.9, C | 150.9, C | 150.4, C | 150.8, C | 150.3, C |
| 4′ | 138.4, C | 138.9, C | 139.0, C | 138.5, C | 138.5, C | 138.4, C |
| 5′ | 136.2, C | 136.7, C | 136.6, C | 136.3, C | 136.0, C | 136.2, C |
| 6′ | 115.3, CH | 115.8, CH | 115.6, CH | 115.3, CH | 115.1, CH | 114.9, CH |
| 7′ | 124.9, CH | 125.4, CH | 125.5, CH | 125.0, CH | 125.0, CH | 124.9, CH |
| 8′ | 75.6, CH | 76.1, CH | 76.1, CH | 75.7, CH | 75.1, CH | 75.6, CH |
| 9′,10′ | 22.0, CH3 | 22.3, CH3 | 22.3, CH3 | 22.0, CH3 | 21.8, CH3 | 22.0, CH3 |
| 1′′ | 164.3, C | 164.8, C | 164.8, C | 164.4, C | 164.1, C | 164.7, C |
| 2′′ | 128.6, C | 129, C | 129.3, C | 128.5, C | 128.3, C | 128.6, C |
| 3′′,7′′ | 128.4, CH | 128.9, CH | 128.9, CH | 128.5, CH | 128.3, CH | 128.4, CH |
| 4′′,6′′ | 118.8, CH | 119.4, CH | 119.3, CH | 118.8, CH | 118.6, CH | 118.8, CH |
| 5′′ | 142.1, C | 142.5, C | 142.6, C | 142.2, C | 142.0, C | 142.1, C |
| 1′′′ | 168.6, C | 168.6, C | 169.2, C | 169.2, C | 168.6, C | 168.6, C |
| 2′′′ | 55.7, CH | 56.0, CH | 56.1, CH | 55.7, CH | 55.6, CH | 55.5, CH |
| 3′′′ | 79.7, CH | 80.2, CH | 80.5, CH | 80.0, CH | 79.9, CH | 79.8, CH |
| 4′′′ | 170.8, C | 171.4, C | 171.3, C | 171.0, C | 171.2, C | 170.8, C |
| 5′′′ | 57.7, CH3 | 58.6, CH3 | 58.1, CH3 | 57.6, CH3 | 57.5, CH3 | 57.6, CH3 |
| 1′′′′ | 165.4, C | 166.2, C | 166.0, C | 166.0, C | 165.5, C | 165.4, C |
| 2′′′′ | 128.9, C | 129.4, C | 129.3, C | 120.3, C | 128.5, C | 128.9, C |
| 3′′′′,7′′′′ | 128.2, CH | 129.0, CH | 129.9, CH | 129.0, CH | 128.4, CH | 128.2, CH |
| 4′′′′,6′′′′ | 119.6, CH | 120.0, CH | 119.7, CH | 112.7, CH | 119.5, CH | 117.6, CH |
| 5′′′′ | 141.7, C | 142.1, C | 143.3, C | 151.8, C | 141.7, C | 141.7, C |
| 1′′′′′ | 164.2, C | 164.6, C | 165.9, C | - | 164.0, C | 164.2, C |
| 2′′′′′ | 140.4, C | 140.7, C | 120.9, C | - | 140.2, C | 140.4, C |
| 3′′′′′,7′′′′′ | 129.3, CH | 129.8, CH | 129.9, CH | - | 129.2, CH | 129.3, CH |
| 4′′′′′,6′′′′′ | 123.5, CH | 124.0, CH | 113.1, CH | - | 123.5, CH | 123.5, CH |
| 5′′′′′ | 149.2, C | 149.7, C | 152.8, C | 149.1, C | 149.2, C |
| Position | 11 | 13 | 1 | |||
|---|---|---|---|---|---|---|
| δC | δH | δC | δH | δC | δH | |
| 1′′′ | 168.6, C | - | 168.7, C | - | 168.6, C | - |
| 2′′′ | 55.7, CH | 4.92, t (8.0) | 54.9, CH | 4.87, brs | 56.0, CH | 5.07, t (8.3) |
| 3′′′ | 79.7, CH | 4.09, d (8.0) | 82.6, CH | 4.27, brs | 80.2, CH | 4.16, d (8.1) |
| 4′′′ | 170.8, C | - | 170.1, C | - | 171.4, C | - |
| 4′′′-NHa | - | 7.47, s | - | - | - | - |
| 4′′′-NHb | - | 7.54, s | - | - | - | - |
| 5′′′ | 57.7, CH3 | 3.31, s | 58.9, CH3 | 3.44, s | 58.6, CH3 | 3.35, s |
| 6′′′-NH | - | 8.46, d (8.1) | - | 8.37, brs | - | 8.72, d (8.5) |
| Position | 10 a | 4 a | 5 b | 6 b | 7 b |
|---|---|---|---|---|---|
| 1 | - | - | - | - | |
| 2 | - | - | - | - | |
| 3 | 7.68, s | 7.68, d (1.6) | 7.59, d (1.7) | 7.58, d (1.5) | 7.55, d (1.7) |
| 4 | - | - | - | - | |
| 5 | - | - | - | - | |
| 6 | 8.50, d (8.8) | 8.53, d (8.4) | 8.36, d (8.3) | 8.54, d (8.3) | 8.44, d (8.4) |
| 7 | 7.69, d (7.4) | 7.71, dd (1.7, 8.4) | 7.63, dd (1.7, 8.3) | 7.60, dd (1.7, 8.4) | 7.61, dd (1.7, 8.4) |
| 8 | 4.80, m | 4.29, q (7.0) | 3.97, s | 4.77, m | 4.21, q (6.9) |
| 9,10 | 1.48, d (8.0) | 1.58, t (7.0) | - | 1.39, d (6.0) | 1.47, t (6.9) |
| 11-NH | - | 10.83, s | 10.96, s | 10.93, s | |
| 1′ | - | - | - | - | |
| 2′ | - | - | - | - | |
| 3′ | - | - | - | - | |
| 3′-OH | - | 11.51, s | 11.32, s | 11.34, s | |
| 4′ | - | - | - | - | |
| 5′ | - | - | - | - | |
| 6′ | 7.80, d (8.6) | 7.83, overlapped | 7.69, d (8.8) | 7.68, d (8.9) | 7.64, d (8.8) |
| 7′ | 7.77, d (9.0) | 7.83, overlapped | 7.81, d (8.8) | 7.80, d (8.9) | 7.81, d (8.8) |
| 8′ | 4.55, m | 4.16, q (7.0) | 4.42, m | 4.02, q (7.0) | 4.36, m |
| 9′,10′ | 1.37, d (6.14) | 1.45, t (7.0) | 1.28, d (6.1) | 1.34, t (7.0) | 1.28, d (6.2) |
| 11′-NH | - | 9.10, s | 9.14, s | 9.25, s | |
| 1′′ | - | - | - | - | |
| 2′′ | - | - | - | - | |
| 3′′,7′′ | 7.78, d (4.8) | 7.77, d (8.7) | 7.70, d (8.7) | 7.71, d (8.7) | 7.77, d (8.6) |
| 4′′,6′′ | 6.88, d (8.5) | 6.79, d (8.7) | 6.64, d (8.6) | 6.64, d (8.7) | 6.81, d (8.2) |
| 5′′ | - | - | - | - |
| Position | 10 a | 4 a | 5 b | 6 b | 7 b |
|---|---|---|---|---|---|
| 1 | 169.5, C | 167.8, C | 167.4, C | 167.5, C | 167.4, C |
| 2 | 127.3, C | 125.9, C | 126.9, C | 126.0, C | 126.3, C |
| 3 | 116.3, CH | 111.8, CH | 111.7, CH | 114.3, CH | 112.6, CH |
| 4 | 148.4, C | 148.0, C | 149.4, C | 146.4, C | 148.3, C |
| 5 | 134.4, C | 132.1, C | 131.9, C | 133.8, C | 132.5, C |
| 6 | 121.2, CH | 119.5, CH | 120.8, CH | 120.0, CH | 120.4, CH |
| 7 | 124.4, CH | 122.7, CH | 123.0, CH | 122.9, CH | 122.8, CH |
| 8 | 73.3, CH | 64.5, CH2 | 56.5, CH3 | 72.1, CH | 64.8, CH2 |
| 9 | 22.3, CH3 | 13.5, CH3 | - | 22.0, CH3 | 14.9, CH3 |
| 10 | 22.3, CH3 | - | - | - | - |
| 1′ | 167.6, C | 165.2, C | 165.0, C | 164.1, C | 164.6, C |
| 2′ | 116.4, C | 114.9, C | 114.8, C | 116.0, C | 116.0, C |
| 3′ | 151.5, C | 150.7, C | 151.4, C | 150.3, C | 151.0, C |
| 4′ | 138.3, C | 137.9, C | 137.9, C | 138.6, C | 138.1, C |
| 5′ | 138.1, C | 136.5, C | 136.9, C | 136.9, C | 137.4, C |
| 6′ | 114.5, CH | 112.9, CH | 113.9, CH | 114.1, CH | 114.5, CH |
| 7′ | 125.5, CH | 124.7, CH | 125.1, CH | 125.9, CH | 125.2, CH |
| 8′ | 77.2, CH | 69.0, CH2 | 75.3, CH | 68.9, CH | 75.8, CH |
| 9′ | 22.7, CH3 | 14.3, CH3 | 22.3, CH3 | 15.6, CH3 | 22.6, CH3 |
| 10′ | 22.7, CH3 | - | 22.3, CH3 | 15.6, CH3 | 22.6, CH3 |
| 1′′ | 167.0, C | 166.4, C | 164.9, C | 165.3, C | 165.1, C |
| 2′′ | 124.1, C | 121.4, C | 120.5, C | 120.2, C | 122.9, C |
| 3′′,7′′ | 130.3, CH | 128.9, CH | 129.5, CH | 129.6, CH | 129.5, CH |
| 4′′,6′′ | 115.1, CH | 113.7, CH | 113.2, CH | 113.2, CH | 115.2, CH |
| 5′′ | 152.9, C | 152.2, C | 152.9, C | 152.9, C | 150.0, C |
| Test Organisms | MIC (μg/mL) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 11 | 12 | 13 | 1 | 2 | 3 | 8 | 9 | Cip * | |
| Staphylococcus aureus RN 4220 | 0.125 | 0.015 | 2 | 4 | 0.25 | >32 | 0.25 | 0.125 | 0.125 |
| MRSA CCARM 3167 | 0.25 | 0.015 | 1 | 4 | 0.5 | >32 | 0.5 | 0.25 | 4 |
| MRSA CCARM 3506 | 0.25 | 0.015 | 1 | 4 | 0.5 | >32 | 0.125 | 0.25 | 2 |
| QRSA CCARM 3505 | 1 | 0.125 | - | 4 | 1 | >32 | 2 | 2 | 128 |
| QRSA CCARM 3519 | 1 | 0.25 | - | 4 | 1 | >32 | 1 | 1 | 128 |
| Streptococcus pneumonia KCTC 5412 | 1 | 0.25 | >16 | 4 | 2 | >32 | 2 | 1 | 0.25 |
| Enterococcus faecalis KCTC 5191 | 0.25 | 0.03 | 4 | 8 | 1 | >32 | 1 | 0.5 | 0.5 |
| Acinetobacter baumannii KCTC 2508 | 2 | 0.125 | 4 | >32 | 4 | >32 | 1 | 0.5 | 0.25 |
| E. coli CCARM 1356 | 2 | 0.125 | 16 | >32 | 1 | >32 | 0.5 | 1 | 64 |
| E. coli KCTC 1682 | 1 | 0.125 | 4 | >32 | 1 | >32 | 0.25 | 0.5 | 0.06 |
| Pseudomonas aeruginosa KCTC 2004 | 4 | 4 | >16 | >32 | >32 | >32 | 4 | 8 | 0.03 |
| Klebsiella pneumoniae KCTC 22057 | 4 | 2 | >16 | >32 | >32 | >32 | >32 | >32 | 0.015 |
| 11 | 12 | 13 | 1 | 2 | 3 | 4 | 8 | 9 | 10 | Cip | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| IC50 | 0.95 | 0.08 | 1.07 | 0.42 | 0.51 | 39.1 | >300 | 0.05 | 0.13 | >300 | 0.30 |
| MIC | 1 | 0.25 | 4 | >32 | 1 | >32 | >32 | 0.25 | 0.25 | >32 | 0.06 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, B.-M.; Minh, N.V.; Choi, H.-Y.; Kim, W.-G. Coralmycin Derivatives with Potent Anti-Gram Negative Activity Produced by the Myxobacteria Corallococcus coralloides M23. Molecules 2019, 24, 1390. https://doi.org/10.3390/molecules24071390
Kim B-M, Minh NV, Choi H-Y, Kim W-G. Coralmycin Derivatives with Potent Anti-Gram Negative Activity Produced by the Myxobacteria Corallococcus coralloides M23. Molecules. 2019; 24(7):1390. https://doi.org/10.3390/molecules24071390
Chicago/Turabian StyleKim, Bo-Min, Nguyen Van Minh, Ha-Young Choi, and Won-Gon Kim. 2019. "Coralmycin Derivatives with Potent Anti-Gram Negative Activity Produced by the Myxobacteria Corallococcus coralloides M23" Molecules 24, no. 7: 1390. https://doi.org/10.3390/molecules24071390
APA StyleKim, B. -M., Minh, N. V., Choi, H. -Y., & Kim, W. -G. (2019). Coralmycin Derivatives with Potent Anti-Gram Negative Activity Produced by the Myxobacteria Corallococcus coralloides M23. Molecules, 24(7), 1390. https://doi.org/10.3390/molecules24071390