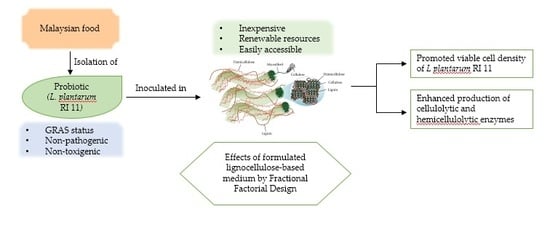
Enhancement of Versatile Extracellular Cellulolytic and Hemicellulolytic Enzyme Productions by Lactobacillus plantarum RI 11 Isolated from Malaysian Food Using Renewable Natural Polymers
Abstract
:1. Introduction
2. Results and Discussion
2.1. Growth Profile of L. plantarum RI 11 in Media Supplemented with Renewable Natural Polymers
2.2. Extracellular Cellulolytic and Hemicellulolytic Enzyme Activities of L. plantarum RI 11
2.3. Fractional Factorial Design for Growth Enhancement of L. plantarum RI 11
2.4. Fractional Factorial Design for Enhancement of Cellulolytic and Hemicellulolytic Enzyme Productions
3. Materials and Methods
3.1. Microorganisms and Maintenance
3.2. Media Formulation for L. plantarum RI 11
3.3. Extracellular Cellulolytic and Hemicellulolytic Enzymes Activities of L. plantarum RI 11
3.4. Protein Concentration Determination
3.5. Cell Viability Determination
3.6. Quantification of Extracellular Cellulolytic and Hemicellulolytic Enzyme Activities
3.7. Fractional Factorial Design for Growth Enhancement and L. plantarum RI 11
3.8. Statistical Analysis
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| GRAS | Generally recognized as safe |
| LAB | Lactic acid bacteria |
| L. plantarum | Lactobacillus plantarum |
| MRS | deMan, Rogosa and Sharpe |
| PKC | Palm kernel cake |
| CFU | Colony forming unit |
| FFD | Fractional factorial design |
| CRMRS | Reconstituted MRS |
| CFS | Cell free supernatant |
| NaCl | Sodium chloride |
| DNS | 3,5-dinitrosalicyclic acid |
| ANOVA | Analysis of variance |
| SEM | Standard error of mean |
References
- Singh, R.M.M. Role of Carbon and Nitrogen Sources in Bacterial Growth and Sporulation. Appl. Microbiol. 1971, 22, 131–132. [Google Scholar] [CrossRef] [Green Version]
- Lochhead, A.G.; Burton, M.O. An Essential Bacterial Growth Factor Produced By Microbial Synthesis. Can. J. Bot. 1953, 31, 7–22. [Google Scholar] [CrossRef]
- De Man, J.C.; Rogosa, M.; Sharpe, M.E. A Medium For The Cultivation Of Lactobacilli. J. Appl. Bacteriol. 1960, 23, 130–135. [Google Scholar] [CrossRef]
- Hayek, S.A. Current Limitations and Challenges with Lactic Acid Bacteria: A Review. Food Nutr. Sci. 2013, 4, 73–87. [Google Scholar] [CrossRef] [Green Version]
- Kirchman, D.L. The contribution of monomers and other low molecular weight compounds to the flux of DOM in aquatic ecosystems. In Aquatic Ecosystems – Dissolved Organic Matter; Findlay, S., Sinsabaugh, R.L., Eds.; Academic Press: New York, NY, USA, 2003; pp. 217–241. [Google Scholar]
- Mikkelsen, D.; Flanagan, B.; Dykes, G.A.; Gidley, M. Influence of different carbon sources on bacterial cellulose production byGluconacetobacter xylinusstrain ATCC 53524. J. Appl. Microbiol. 2009, 107, 576–583. [Google Scholar] [CrossRef] [PubMed]
- Ravindran, R.; Jaiswal, A.K. Microbial Enzyme Production Using Lignocellulosic Food Industry Wastes as Feedstock: A Review. Bioengineering 2016, 3, 30. [Google Scholar] [CrossRef] [Green Version]
- Lim, Y.H.; Foo, H.L.; Loh, T.C.; Mohamad, R.; Rahim, R.A. Rapid Evaluation and Optimization of Medium Components Governing Tryptophan Production by Pediococcus acidilactici TP-6 Isolated from Malaysian Food via Statistical Approaches. Molecules 2020, 25, 779. [Google Scholar] [CrossRef] [Green Version]
- Lim, Y.H.; Foo, H.L.; Loh, T.C.; Mohamad, R.; Rahim, R.A.; Idrus, Z. Optimized medium via statistical approach enhanced threonine production by Pediococcus pentosaceus TL-3 isolated from Malaysian food. Microb. Cell Factories 2019, 18, 125. [Google Scholar] [CrossRef]
- Djekrif-Dakhmouche, S.; Gheribi-Aoulmi, Z.; Meraihi, Z.; Bennamoun, L. Application of a statistical design to the optimization of culture medium for α-amylase production by Aspergillus niger ATCC 16404 grown on orange waste powder. J. Food Eng. 2006, 73, 190–197. [Google Scholar] [CrossRef]
- Guo, F.; Li, X.; Zhao, J.; Li, G.; Gao, P.; Han, X. Optimizing Culture Conditions by Statistical Approach to Enhance Production of Pectinase from Bacillus sp. Y1. BioMed Res. Int. 2019, 2019, 8146948. [Google Scholar] [CrossRef] [Green Version]
- Abdullah, N.; Sulaim, F. The Oil Palm Wastes in Malaysia. In Biomass Now - Sustainable Growth and Use; IntechOpen: London, UK, 2013. [Google Scholar]
- Hengniran, P. Future Potential of Forest and Agriculture Residues for the Energy Production in Thailand-Strategies for a Better Utilization. Ph.D. Thesis, University of Hamburg, Hamburg, Germany, 2010. [Google Scholar]
- Pariza, M.W.; Johnson, E.A. Evaluating the Safety of Microbial Enzyme Preparations Used in Food Processing: Update for a New Century. Regul. Toxicol. Pharmacol. 2001, 33, 173–186. [Google Scholar] [CrossRef] [PubMed]
- Maki, M. The prospects of cellulase-producing bacteria for the bioconversion of lignocellulosic biomass. Int. J. Boil. Sci. 2009, 5, 500–516. [Google Scholar] [CrossRef] [PubMed]
- Somerville, C.; Youngs, H.; Taylor, C.; Davis, S.C.; Long, S. Feedstocks for Lignocellulosic Biofuels. Science 2010, 329, 790–792. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brodeur, G.; Yau, E.; Badal, K.; Collier, J.; Ramachandran, K.B.; Ramakrishnan, S. Chemical and Physicochemical Pretreatment of Lignocellulosic Biomass: A Review. Enzym. Res. 2011, 2011, 1–17. [Google Scholar] [CrossRef]
- Luterbacher, J.S.; Rand, J.M.; Alonso, D.M.; Han, J.; Youngquist, J.T.; Maravelias, C.T.; Pfleger, B.F.; Dumesic, J.A. Nonenzymatic Sugar Production from Biomass Using Biomass-Derived -Valerolactone. Science 2014, 343, 277–280. [Google Scholar] [CrossRef] [PubMed]
- Isikgor, F.H.; Becer, C.R. Lignocellulosic biomass: a sustainable platform for the production of bio-based chemicals and polymers. Polym. Chem. 2015, 6, 4497–4559. [Google Scholar] [CrossRef] [Green Version]
- Pu, Y.; Hu, F.; Huang, F.; Davison, B.H.; Ragauskas, A. Assessing the molecular structure basis for biomass recalcitrance during dilute acid and hydrothermal pretreatments. Biotechnol. Biofuels 2013, 6, 15. [Google Scholar] [CrossRef] [Green Version]
- Ponnambalam, A.S.; Deepthi, R.S.; Ghosh, A.R. Qualitative display and measurement of enzyme activity of isolated cellulolytic bacteria. Biotechnol. Bioinf. Bioeng. 2011, 1, 33–37. [Google Scholar]
- Martinez, D.; Berka, R.M.; Henrissat, B.; Saloheimo, M.; Arvas, M.; Baker, S.E.; Chapman, J.; Chertkov, O.; Coutinho, P.M.; Cullen, D.; et al. Genome sequencing and analysis of the biomass-degrading fungus Trichoderma reesei (syn. Hypocrea jecorina). Nat. Biotechnol. 2008, 26, 553–560. [Google Scholar] [CrossRef] [Green Version]
- Peng, X.; Qiao, W.; Mi, S.; Jia, X.; Su, H.; Han, Y. Characterization of hemicellulase and cellulase from the extremely thermophilic bacterium Caldicellulosiruptor owensensis and their potential application for bioconversion of lignocellulosic biomass without pretreatment. Biotechnol. Biofuels 2015, 8, 131. [Google Scholar] [CrossRef] [Green Version]
- Han, S.O.; Cho, H.-Y.; Yukawa, H.; Inui, M.; Doi, R.H. Regulation of Expression of Cellulosomes and Noncellulosomal (Hemi)Cellulolytic Enzymes in Clostridium cellulovorans during Growth on Different Carbon Sources. J. Bacteriol. 2004, 186, 4218–4227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sánchez, C. Lignocellulosic residues: Biodegradation and bioconversion by fungi. Biotechnol. Adv. 2009, 27, 185–194. [Google Scholar] [CrossRef] [PubMed]
- Tejirian, A.; Xu, F. Inhibition of enzymatic cellulolysis by phenolic compounds. Enzym. Microb. Technol. 2011, 48, 239–247. [Google Scholar] [CrossRef] [PubMed]
- Dashtban, M. Fungal Bioconversion of Lignocellulosic Residues; Opportunities & Perspectives. Int. J. Boil. Sci. 2009, 5, 578–595. [Google Scholar]
- Tshikantwa, T.S.; Ullah, M.W.; He, F.; Yang, G. Current Trends and Potential Applications of Microbial Interactions for Human Welfare. Front. Microbiol. 2018, 9, 1156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Generally Regarded As Safe (GRAS). Available online: https://www.fda.gov/food/food-ingredients-packaging/generally-recognized-safe-gras (accessed on 22 February 2020).
- Sewalt, V.; Shanahan, D.; Gregg, L.; La Marta, J.; Carrillo, R. The Generally Recognized as Safe (GRAS) Process for Industrial Microbial Enzymes. Ind. Biotechnol. 2016, 12, 295–302. [Google Scholar] [CrossRef] [Green Version]
- Åvall-Jääskeläinen, S.; Palva, A. Lactobacillus surface layers and their applications. FEMS Microbiol. Rev. 2005, 29, 511–529. [Google Scholar] [CrossRef] [Green Version]
- Choi, S.S.; Kang, B.Y.; Chung, M.J.; Kim, S.D.; Park, S.H.; Kim, J.S.; Kang, C.Y.; Ha, N.J. Safety assessment of potential lactic acid bacteria Bifidobacterium longum SPM1205 isolated from healthy Koreans. J. Microbiol. 2005, 43, 493–498. [Google Scholar]
- Gueimonde, M.; Frias, R.; Ouwehand, A. Assuring the continued safety of lactic acid bacteria used as probiotics. Boilogia 2006, 61, 755–760. [Google Scholar] [CrossRef]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef] [Green Version]
- Foo, H.L.; Loh, T.C.; Law, F.L.; Lim, Y.S.; Kufli, C.N.; Rusul, G. Effect of feeding Lactobacillus plantarum I-UL4 isolated from Malaysian Tempeh on growth performance, faecal flora and lactic acid bacteria and plasma cholesterol concentrations in postweaning rats. Food Sci. Biotechnol. 2003, 12, 403–408. [Google Scholar]
- Moghadam, M.S.; Foo, H.L.; Leow, T.C.; Rahim, R.A.; Loh, T.C. Novel bacteriocinogenic Lactobacillus plantarum strains and their differentiation by sequence analysis of 16S rDNA, 16S-5S intergenic spacer region and randomly amplied polymorphic DNA analysis. Food Technol. Biotechnol. 2010, 48, 476–483. [Google Scholar]
- Kareem, K.Y.; Foo, H.L.; Loh, T.C.; Foong, O.M.; Samsudin, A.A. Inhibitory activity of postbiotic produced by strains of Lactobacillus plantarum using reconstituted media supplemented with inulin. Gut Pathogens 2014, 6, 23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loh, T.C.; Thanh, N.; Foo, H.L.; Hair-Bejo, M. Effects of feeding metabolite combinations from lactobacillus plantarum on plasma and breast meat lipids in Broiler Chickens. Braz. J. Poultry Sci. 2013, 15, 307–316. [Google Scholar] [CrossRef] [Green Version]
- Lim, Y.H.; Foo, H.L.; Loh, T.C.; Mohamad, R.; Abdullah, N. Comparative studies of versatile extracellular proteolytic activities of lactic acid bacteria and their potential for extracellular amino acid productions as feed supplements. J. Anim. Sci. Biotechnol. 2019, 10, 15. [Google Scholar] [CrossRef] [Green Version]
- Kareem, K.Y.; Loh, T.C.; Foo, H.L.; Akit, H.; Samsudin, A.A. Effects of dietary postbiotic and inulin on growth performance, IGF1 and GHR mRNA expression, faecal microbiota and volatile fatty acids in broilers. BMC Veter- Res. 2016, 12, 163. [Google Scholar] [CrossRef] [Green Version]
- Kareem, K.Y.; Loh, T.C.; Foo, H.L.; Asmara, A.S.; Akit, H.; Abdulla, N.R.; Ooi, M.F. Carcass, meat and bone quality of broiler chickens fed with postbiotic and prebiotic combinations. Int. J. Probiotics Prebiotics. 2015, 10, 23. [Google Scholar]
- Tarraran, L.; Mazzoli, R. Alternative strategies for lignocellulose fermentation through lactic acid bacteria: the state of the art and perspectives. FEMS Microbiol. Lett. 2018, 365. [Google Scholar] [CrossRef]
- Toe, C.J.; Foo, H.L.; Loh, T.C.; Mohamad, R.; Rahim, R.A.; Zulkifli, I. Extracellular Proteolytic Activity and Amino Acid Production by Lactic Acid Bacteria Isolated from Malaysian Foods. Int. J. Mol. Sci. 2019, 20, 1777. [Google Scholar] [CrossRef] [Green Version]
- Heenan, C.; Adams, M.; Hosken, R.; Fleet, G. Growth Medium for Culturing Probiotic Bacteria for Applications in Vegetarian Food Products. LWT 2002, 35, 171–176. [Google Scholar] [CrossRef]
- Douillard, F.P.; De Vos, W.M. Functional genomics of lactic acid bacteria: from food to health. In Microbial Cell Factories; BioMed Central: London, UK, 2014; Volume 13, p. S8. [Google Scholar]
- Boguta, A.; Bringel, F.; Martinussen, J.; Jensen, P.R. Screening of lactic acid bacteria for their potential as microbial cell factories for bioconversion of lignocellulosic feedstocks. Microb. Cell Factories 2014, 13, 97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taskila, S.; Ojamo, H. The Current Status and Future Expectations in Industrial Production of Lactic Acid by Lactic Acid Bacteria. In Lactic Acid Bacteria-R & D for Food, Health and Livestock Purposes; IntechOpen: London, UK, 2013. [Google Scholar]
- Lee, F.; Wan, S.; Foo, H.L.; Loh, T.C.; Mohamad, R.; Rahim, R.A.; Zulkifli, I. Comparative Study of Extracellular Proteolytic, Cellulolytic, and Hemicellulolytic Enzyme Activities and Biotransformation of Palm Kernel Cake Biomass by Lactic Acid Bacteria Isolated from Malaysian Foods. Int. J. Mol. Sci. 2019, 20, 4979. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cuenca, A.R.; Suárez, M.J.V.; Mateos-Aparicio, I. Soybean seeds and its by-product okara as sources of dietary fibre. Measurement by AOAC and Englyst methods. Food Chem. 2008, 108, 1099–1105. [Google Scholar] [CrossRef]
- Lu, F.; Liu, Y.; Li, B. Okara dietary fiber and hypoglycemic effect of okara foods. Bioact. Carbohydr. Diet. Fibre 2013, 2, 126–132. [Google Scholar] [CrossRef]
- Shikha; Sharan, A.; Darmwal, N. Improved production of alkaline protease from a mutant of alkalophilic Bacillus pantotheneticus using molasses as a substrate. Bioresour. Technol. 2007, 98, 881–885. [Google Scholar] [CrossRef] [PubMed]
- Coelho, L.F.; De Lima, C.J.B.; Rodovalho, C.D.M.; Bernardo, M.P.; Contiero, J. Lactic acid production by new Lactobacillus plantarum LMISM6 grown in molasses: optimization of medium composition. Braz. J. Chem. Eng. 2011, 28, 27–36. [Google Scholar] [CrossRef] [Green Version]
- Razmovski, R.; Vučurović, V. Ethanol production from sugar beet molasses by S. cerevisiae entrapped in an alginate–maize stem ground tissue matrix. Enzym. Microb. Technol. 2011, 48, 378–385. [Google Scholar] [CrossRef]
- El-Gendy, N.S.; Madian, H.R.; Abu Amr, S.S. Design and Optimization of a Process for Sugarcane Molasses Fermentation by Saccharomyces cerevisiae Using Response Surface Methodology. Int. J. Microbiol. 2013, 2013, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Hamouda, H.I.; Nassar, H.; Madian, H.R.; El-Sayed, M.H.; El-Ghamry, A.A.; El-Gendy, N.S. Isolation of fermentative microbial isolates from sugar cane and beet molasses and evaluation for enhanced production of bioethanol. Energy Sour. 2016, 38, 2170–2180. [Google Scholar] [CrossRef]
- Pholsen, S.; Khota, W.; Pang, H.; Higgs, D.; Cai, Y. Characterization and application of lactic acid bacteria for tropical silage preparation. Anim. Sci. J. 2016, 87, 1202–1211. [Google Scholar] [CrossRef]
- Jin, S.; Chen, H. Structural properties and enzymatic hydrolysis of rice straw. Process. Biochem. 2006, 41, 1261–1264. [Google Scholar] [CrossRef]
- Zajšek, K.; Goršek, A.; Kolar, M. Cultivating conditions effects on kefiran production by the mixed culture of lactic acid bacteria imbedded within kefir grains. Food Chem. 2013, 139, 970–977. [Google Scholar] [CrossRef] [PubMed]
- Deutscher, J. The mechanisms of carbon catabolite repression in bacteria. Curr. Opin. Microbiol. 2008, 11, 87–93. [Google Scholar] [CrossRef] [PubMed]
- Da Silva-Maia, J.K.; Cazarin, C.B.B.; Júnior, S.B.; Augusto, F.; Junior, M.R.M. Passion fruit (Passiflora edulis) peel increases colonic production of short-chain fatty acids in Wistar rats. LWT 2014, 59, 1252–1257. [Google Scholar] [CrossRef]
- Ravindran, R.; Hassan, S.; Williams, G.; Jaiswal, A.K. A Review on Bioconversion of Agro-Industrial Wastes to Industrially Important Enzymes. Bioengineering 2018, 5, 93. [Google Scholar] [CrossRef] [Green Version]
- Horn, S.J.; Vaaje-Kolstad, G.; Westereng, B.; Eijsink, V.G.H. Novel enzymes for the degradation of cellulose. Biotechnol. Biofuels 2012, 5, 45. [Google Scholar] [CrossRef] [Green Version]
- Horn, S.J.; Sikorski, P.; Cederkvist, J.B.; Vaaje-Kolstad, G.; Sørlie, M.; Synstad, B.; Vriend, G.; Vårum, K.M.; Eijsink, V.G.H. Costs and benefits of processivity in enzymatic degradation of recalcitrant polysaccharides. Proc. Natl. Acad. Sci. USA 2006, 103, 18089–18094. [Google Scholar] [CrossRef] [Green Version]
- Payne, C.; Bomble, Y.J.; Taylor, C.B.; McCabe, C.; Himmel, M.E.; Crowley, M.F.; Beckham, G.T. Multiple Functions of Aromatic-Carbohydrate Interactions in a Processive Cellulase Examined with Molecular Simulation. J. Boil. Chem. 2011, 286, 41028–41035. [Google Scholar] [CrossRef] [Green Version]
- Mahajan, P.M.; Desai, K.M.; Lele, S.S. Production of Cell Membrane-Bound α- and β-Glucosidase by Lactobacillus acidophilus. Food Bioprocess Technol. 2010, 5, 706–718. [Google Scholar] [CrossRef]
- Ghose, T.K. Measurement of cellulase activities. Pure Appl. Chem. 1987, 59, 257–268. [Google Scholar] [CrossRef]
- Miller, G.L. Use of Dinitrosalicylic Acid Reagent for Determination of Reducing Sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Nidetzky, B.; Steiner, W.; Hayn, M.; Claeyssens, M. Cellulose hydrolysis by the cellulases from Trichoderma reesei: a new model for synergistic interaction. Biochem. J. 1994, 298, 705–710. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eriksson, K.-E. Enzyme mechanisms involved in cellulose hydrolysis by the rot fungusSporotrichum pulverulentum. Biotechnol. Bioeng. 1978, 20, 317–332. [Google Scholar] [CrossRef]
- Lin, L.; Kan, X.; Yan, H.; Wang, D. Characterization of extracellular cellulose-degrading enzymes from Bacillus thuringiensis strains. Electron. J. Biotechnol. 2012, 15, 2. [Google Scholar] [CrossRef]
- Spano, G.; Rinaldi, A.; Ugliano, M.; Moio, L.; Beneduce, L.; Massa, S. A beta-glucosidase gene isolated from wine Lactobacillus plantarum is regulated by abiotic stresses. J. Appl. Microbiol. 2005, 98, 855–861. [Google Scholar] [CrossRef] [PubMed]
- Zanoelo, F.; Polizeli, M.D.; Terenzi, H.F.; Jorge, J.A. β-Glucosidase activity from the thermophilic fungus Scytalidium thermophilum is stimulated by glucose and xylose. FEMS Microbiol. Lett. 2004, 240, 137–143. [Google Scholar] [CrossRef] [Green Version]
- Murashima, K.; Kosugi, A.; Doi, R.H. Synergistic Effects on Crystalline Cellulose Degradation between Cellulosomal Cellulases from Clostridium cellulovorans. J. Bacteriol. 2002, 184, 5088–5095. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.; Lee, M.-H.; Lee, E.-S.; Nam, Y.-D.; Seo, D.-H. Characterization of mannanase from Bacillus sp., a novel Codium fragile cell wall-degrading bacterium. Food Sci. Biotechnol. 2017, 27, 115–122. [Google Scholar] [CrossRef]
- El-Sharouny, E.E.; El-Toukhy, N.M.; El-Sersy, N.A.; El-Gayar, A.A.E.-A. Optimization and purification of mannanase produced by an alkaliphilic-thermotolerant Bacillus cereus N1 isolated from Bani Salama Lake in Wadi El-Natron. Biotechnol. Biotechnol. Equip. 2015, 29, 315–323. [Google Scholar] [CrossRef]
- Rasul, F.; Afroz, A.; Rashid, U.; Mehmood, S.; Sughra, K.; Zeeshan, N. Screening and characterization of cellulase producing bacteria from soil and waste (molasses) of sugar industry. Int. J. Biosci. 2015, 6, 230–236. [Google Scholar]
- Sachslehner, A.; Nidetzky, B.; Kulbe, K.D.; Haltrich, D. Induction of Mannanase, Xylanase, and Endoglucanase Activities in Sclerotium rolfsii. Appl. Environ. Microbiol. 1998, 64, 594–600. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allcock, E.R.; Woods, D.R. Carboxymethyl cellulase and cellobiase production by Clostridium acetobutylicum in an industrial fermentation medium. Appl. Environ. Microbiol. 1981, 41, 539–541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cho, S.J. Isolation and characterization of mannanase producing Bacillus amyloliquefaciens CS47 from horse feces. J. Life Sci. 2009, 19, 1724–1730. [Google Scholar]
- Gunst, R.F.; Mason, R.L. Fractional factorial design. Wiley Interdiscip. Rev. Comput. Stat. 2009, 1, 234–244. [Google Scholar] [CrossRef]
- Das, A.K.; Dewanjee, S. Optimization of Extraction Using Mathematical Models and Computation. In Computational Phytochemistry; Elsevier BV: Amsterdam, The Netherlands, 2018; pp. 75–106. [Google Scholar]
- Rahman, A.S. Effects of nanofibers on properties of geopolymer composites. In Nanotechnology in Eco-Efficient Construction; Elsevier BV: Amsterdam, The Netherlands, 2019; pp. 123–140. [Google Scholar]
- Watson, S.P.; Clements, M.O.; Foster, S.J. Characterization of the Starvation-Survival Response of Staphylococcus aureus. J. Bacteriol. 1998, 180, 1750–1758. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Xia, K.; Yang, X.; Tang, C. Growth strategy of microbes on mixed carbon sources. Nat. Commun. 2019, 10, 1279. [Google Scholar] [CrossRef] [PubMed]
- Stuelke, J.; Hillen, W. Carbon catabolite repression in bacteria. Curr. Opin. Microbiol. 1999, 2, 195–201. [Google Scholar] [CrossRef]
- Jia, X.; Chen, M.; Wan, J.-B.; Su, H.; He, C. Review on the extraction, characterization and application of soybean polysaccharide. RSC Adv. 2015, 5, 73525–73534. [Google Scholar] [CrossRef]
- El Sharkawi, H.M. Effect of Nitrogen Sources on Microbial Biomass Nitrogen under Different Soil Types. ISRN Soil Sci. 2012, 2012, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Li, B.; Qiao, M.; Lu, F. Composition, Nutrition, and Utilization of Okara (Soybean Residue). Food Rev. Int. 2012, 28, 231–252. [Google Scholar] [CrossRef]
- Duc, C.; Pradal, M.; Sanchez, I.; Noble, J.; Tesnière, C.; Blondin, B. A set of nutrient limitations trigger yeast cell death in a nitrogen-dependent manner during wine alcoholic fermentation. PLoS ONE 2017, 12, e0184838. [Google Scholar] [CrossRef] [PubMed]
- Costa, E.; Teixidó, N.; Usall, J.; Atares, E.; Viñas, I. The effect of nitrogen and carbon sources on growth of the biocontrol agent Pantoea agglomerans strain CPA-2. Lett. Appl. Microbiol. 2002, 35, 117–120. [Google Scholar] [CrossRef] [PubMed]
- Brückner, R.; Titgemeyer, F. Carbon catabolite repression in bacteria: choice of the carbon source and autoregulatory limitation of sugar utilization. FEMS Microbiol. Lett. 2002, 209, 141–148. [Google Scholar] [CrossRef]
- Dashtban, M.; Buchkowski, R.; Qin, W. Effect of different carbon sources on cellulase production by Hypocrea jecorina (Trichoderma reesei) strains. Int. J. Biochem. Mol. Boil. 2011, 2, 274–286. [Google Scholar]
- Stanbury, P.F.; Whitaker, A.; Hall, S.J. Principles of Fermentation Technology; Elsevier: Amsterdam, The Netherlands, 2013. [Google Scholar]
- Egli, T. Microbial growth and physiology: a call for better craftsmanship. Front. Microbiol. 2015, 6, 287. [Google Scholar] [CrossRef] [PubMed]
- Dou, T.-Y.; Chen, J.; Hao, Y.-F.; Qi, X. Effects of Different Carbon Sources on Enzyme Production and Ultrastructure of Cellulosimicrobium cellulans. Curr. Microbiol. 2019, 76, 355–360. [Google Scholar] [CrossRef] [PubMed]
- Ahirwar, S.; Soni, H.; Rawat, H.K.; Prajapati, B.P.; Kango, N. Experimental design of response surface methodology used for utilisation of palm kernel cake as solid substrate for optimised production of fungal mannanase. Mycology 2016, 7, 143–153. [Google Scholar] [CrossRef] [Green Version]
- Lloyd, D.K.; Bergum, J. Application of quality by design (QbD) to the development and validation of analytical methods. In Specification of Drug Substances and Products; Elsevier: Amsterdam, The Netherlands, 2014; pp. 29–72. [Google Scholar]
- A Mooney, C.; Mansfield, S.D.; Beatson, R.P.; Saddler, J.N. The effect of fiber characteristics on hydrolysis and cellulase accessibility to softwood substrates. Enzym. Microb. Technol. 1999, 25, 644–650. [Google Scholar] [CrossRef]
- Gao, D.; Chundawat, S.P.S.; Krishnan, C.; Balan, V.; Dale, B.E. Mixture optimization of six core glycosyl hydrolases for maximizing saccharification of ammonia fiber expansion (AFEX) pretreated corn stover. Bioresour. Technol. 2010, 101, 2770–2781. [Google Scholar] [CrossRef]
- Kuhad, R.C.; Gupta, R.; Singh, A. Microbial Cellulases and Their Industrial Applications. Enzym. Res. 2011, 2011, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Gao, D.; Uppugundla, N.; Chundawat, S.P.S.; Yu, X.; Hermanson, S.; Gowda, K.; Brumm, P.; Mead, D.; Balan, V.; Dale, B.E. Hemicellulases and auxiliary enzymes for improved conversion of lignocellulosic biomass to monosaccharides. Biotechnol. Biofuels 2011, 4, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arotupin, D.; Olaniyi, O.O. Screening and Identification of Mannanase-Producing Fungi Isolated from Selected Agricultural Wastes. Br. Microbiol. Res. J. 2013, 3, 635–644. [Google Scholar] [CrossRef]
- Zakaria, M.M.; Ashiuchi, M.; Yamamoto, S.; Yagi, T. Optimization for β-Mannanase Production of a Psychrophilic Bacterium, Flavobacteriumsp. Biosci. Biotechnol. Biochem. 1998, 62, 655–660. [Google Scholar] [CrossRef] [PubMed]
- Aziz, N.F.H.A.; Abbasiliasi, S.; Ng, H.S.; Phapugrangkul, P.; Abu Bakar, M.H.; Tam, Y.J.; Tan, J.S. Purification of β -mannanase derived from Bacillus subtilis ATCC 11774 using ionic liquid as adjuvant in aqueous two-phase system. J. Chromatogr. B 2017, 1055, 104–112. [Google Scholar] [CrossRef] [PubMed]
- Anderson, R.K.I.; Jayaraman, K. Influence of carbon and nitrogen sources on the growth and sporulation of Bacillus thuringiensis var Galleriae for biopesticide production. Chem. Biochem. Eng. Q. 2003, 17, 225–232. [Google Scholar]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Yin, L.J.; Lin, H.H.; Xiao, Z.R. Purification and characterization of a cellulase from Bacillus subtilis YJ1. J. Mar. Sci. Tech. 2010, 18, 466–471. [Google Scholar]
Sample Availability: Samples of the compounds are not available from the authors. |
















| Day | Viable Cell Density(Log CFU/mL) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| F1 | F2 | F3 | F4 | F5 | F6 | F7 | F8 | F9 | |
| 0 | 7.99 ± 0.04Ad | 8.53 ± 0.00Db | 9.43 ± 0.21Ba | 9.25 ± 0.29Ea | 9.40 ± 0.19Da | 9.25 ± 0.15Fa | 8.48 ± 0.04Cbc | 8.28 ± 0.03Dbcd | 7.91 ± 0.10Dd |
| 1 | 7.85 ± 0.30Ae | 9.62 ± 0.26Ee | 9.70 ± 0.04Ba | 9.74 ± 0.03Da | 9.63 ± 0.11Dab | 9.67 ± 0.04DEa | 8.43 ± 0.02Cd | 9.50 ± 0.04Aab | 9.56 ± 0.09Cab |
| 2 | 6.56 ± 0.14Bj | 9.73 ± 0.03Bd | 9.73 ± 0.02Bd | 9.73 ± 0.02Dd | 9.64 ± 0.04Dd | 9.76 ± 0.01Dd | 7.57 ± 0.08Di | 9.22 ± 0.04Be | 10.50 ± 0.01Ac |
| 3 | 6.40 ± 0.10Bc | 10.05 ± 0.07Bc | 10.14 ± 0.09Aab | 10.45 ± 0.03BCab | 10.46 ± 0.05Cab | 10.73 ± 0.01Ba | 9.05 ± 0.19Babc | 8.13 ± 0.14DEabc | 9.41 ± 0.12Cab |
| 4 | 5.92 ± 0.09Ck | 10.61 ± 0.23Ac | 9.20 ± 0.01Cef | 11.76 ± 0.01Aa | 11.76 ± 0.01Aa | 10.99 ± 0.05Bb | 8.68 ± 0.02Chi | 8.74 ± 0.10Cghi | 9.32 ± 0.07Ce |
| 5 | 5.30 ± 0.00Dj | 9.25 ± 0.18Cg | 10.42 ± 0.09Ad | 10.74 ± 0.01Bc | 11.69 ± 0.04Aa | 11.43 ± 0.12Ab | 9.49 ± 0.09Agf | 9.43 ± 0.04ABgf | 10.54 ± 0.07Acd |
| 6 | 0 ± 0Ej | 9.77 ± 0.08Bc | 9.26 ± 0.16Cde | 10.34 ± 0.06BCb | 11.10 ± 0.07Da | 9.43 ± 0.13EFd | 7.46 ± 0.06Di | 7.94 ± 0.11Eh | 10.42 ± 0.06Ab |
| 7 | 0 ± 0Eh | 10.20 ± 0.17ABb | 10.19 ± 0.16Ab | 10.14 ± 0.16Dbc | 11.29 ± 0.07Ba | 10.05 ± 0.16Cbc | 7.28 ± 0.17Dg | 7.90 ± 0.10Ef | 10.01 ± 0.11Bbc |
| F10 | F11 | F12 | F13 | F14 | F15 | F16 | MRS | CRMRS | |
| 0 | 8.08 ± 0.17Dcd | 8.28 ± 0.04BCbcd | 8.12 ± 0.14CDbcd | 8.20 ± 0.01Dbcd | 8.13 ± 0.12Cbcd | 8.29 ± 0.16Ebcd | 8.19 ± 0.05Ebcd | 9.13 ± 0.03Ea | 9.01 ± 0.19Ca |
| 1 | 9.67 ± 0.08Aa | 9.29 ± 0.08Abc | 7.89 ± 0.08De | 9.04 ± 0.09Ac | 8.23 ± 0.10Cd | 9.69 ± 0.04Ca | 9.73 ± 0.02Ca | 9.65 ± 0.00Dab | 9.62 ± 0.04Bab |
| 2 | 9.68 ± 0.06Ad | 9.14 ± 0.02Ae | 7.87 ± 0.17Dh | 9.00 ± 0.02Af | 8.26 ± 0.08Cg | 10.76 ± 0.01Aa | 10.56 ± 0.02Ac | 10.73 ± 0.01Aab | 9.70 ± 0.02ABd |
| 3 | 8.45 ± 0.11Cabc | 8.07 ± 0.03Cabc | 7.86 ± 0.05Dbc | 8.44 ± 0.07Cabc | 8.24 ± 0.02Cabc | 10.76 ± 0.01Aa | 8.42 ± 0.05Eabc | 10.10 ± 0.08Cab | 9.99 ± 0.16Aab |
| 4 | 9.15 ± 0.02Bef | 8.21 ± 0.08BCj | 8.52 ± 0.16ABi | 8.60 ± 0.05BCi | 8.59 ± 0.14Bi | 9.99 ± 0.08Bd | 8.92 ± 0.18Dfgh | 10.36 ± 0.05Bc | 8.99 ± 0.07Cfg |
| 5 | 9.40 ± 0.08Bgf | 8.36 ± 0.09Bi | 8.73 ± 0.02Ah | 8.77 ± 0.01Bh | 8.32 ± 0.04Ci | 10.16 ± 0.08Be | 9.97 ± 0.05BCe | 10.62 ± 0.08Acd | 9.50 ± 0.08Bf |
| 6 | 9.31 ± 0.06Bde | 9.11 ± 0.08Aef | 8.40 ± 0.10ABCg | 8.20 ± 0.06Dgh | 8.95 ± 0.15Af | 9.31 ± 0.07Dde | 10.16 ± 0.11Bb | 9.43 ± 0.05Dd | 8.46 ± 0.06Dg |
| 7 | 9.30 ± 0.01Bd | 7.66 ± 0.14Df | 8.35 ± 0.07BCe | 7.89 ± 0.10Ef | 9.08 ± 0.11Ad | 9.36 ± 0.06Dd | 9.78 ± 0.08Cc | 9.04 ± 0.20Ed | 8.54 ± 0.04De |
| Medium | C Source | Concentration (g/L) | N Source | Concentration (g/L) |
|---|---|---|---|---|
| M1 | Rice straw | 15.46 | Yeast extract | 28.34 |
| M2 | Rice straw | 15.46 | Soybean pulp | 51.54 |
| M3 | PKC | 11.86 | Yeast extract | 28.34 |
| M4 | PKC | 11.86 | Soybean pulp | 51.54 |
| M5 | Molasses | 25.09 | Yeast extract | 28.34 |
| M6 | Molasses | 25.09 | Soybean pulp | 51.54 |
| Control | Glucose | 20.00 | Yeast extract | 28.34 |
| Component | Carbon Content (%) | Nitrogen Content (%) | |
|---|---|---|---|
| Renewable natural polymers | PKC | 31.00 | - |
| Molasses | 14.65 | - | |
| Rice straw | 23.77 | - | |
| Soybean pulp | - | 35.77 | |
| Reconstituted MRS medium | Glucose | 18.38 | - |
| Yeast extract | - | 65.05 |
| Run | Factor | |||||
|---|---|---|---|---|---|---|
| A | B | C | D | E | F | |
| F1 | −1 | −1 | −1 | −1 | −1 | −1 |
| F2 | 1 | −1 | −1 | −1 | 1 | −1 |
| F3 | −1 | 1 | −1 | −1 | 1 | 1 |
| F4 | 1 | 1 | −1 | −1 | −1 | 1 |
| F5 | −1 | −1 | 1 | −1 | 1 | 1 |
| F6 | 1 | −1 | 1 | −1 | −1 | 1 |
| F7 | −1 | 1 | 1 | −1 | −1 | −1 |
| F8 | 1 | 1 | 1 | −1 | 1 | −1 |
| F9 | −1 | −1 | −1 | 1 | −1 | 1 |
| F10 | 1 | −1 | −1 | 1 | 1 | 1 |
| F11 | −1 | 1 | −1 | 1 | 1 | −1 |
| F12 | 1 | 1 | −1 | 1 | −1 | −1 |
| F13 | −1 | −1 | 1 | 1 | 1 | −1 |
| F14 | 1 | −1 | 1 | 1 | −1 | −1 |
| F15 | −1 | 1 | 1 | 1 | −1 | 1 |
| F16 | 1 | 1 | 1 | 1 | 1 | 1 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mohamad Zabidi, N.A.; Foo, H.L.; Loh, T.C.; Mohamad, R.; Abdul Rahim, R. Enhancement of Versatile Extracellular Cellulolytic and Hemicellulolytic Enzyme Productions by Lactobacillus plantarum RI 11 Isolated from Malaysian Food Using Renewable Natural Polymers. Molecules 2020, 25, 2607. https://doi.org/10.3390/molecules25112607
Mohamad Zabidi NA, Foo HL, Loh TC, Mohamad R, Abdul Rahim R. Enhancement of Versatile Extracellular Cellulolytic and Hemicellulolytic Enzyme Productions by Lactobacillus plantarum RI 11 Isolated from Malaysian Food Using Renewable Natural Polymers. Molecules. 2020; 25(11):2607. https://doi.org/10.3390/molecules25112607
Chicago/Turabian StyleMohamad Zabidi, Nursyafiqah A., Hooi Ling Foo, Teck Chwen Loh, Rosfarizan Mohamad, and Raha Abdul Rahim. 2020. "Enhancement of Versatile Extracellular Cellulolytic and Hemicellulolytic Enzyme Productions by Lactobacillus plantarum RI 11 Isolated from Malaysian Food Using Renewable Natural Polymers" Molecules 25, no. 11: 2607. https://doi.org/10.3390/molecules25112607
APA StyleMohamad Zabidi, N. A., Foo, H. L., Loh, T. C., Mohamad, R., & Abdul Rahim, R. (2020). Enhancement of Versatile Extracellular Cellulolytic and Hemicellulolytic Enzyme Productions by Lactobacillus plantarum RI 11 Isolated from Malaysian Food Using Renewable Natural Polymers. Molecules, 25(11), 2607. https://doi.org/10.3390/molecules25112607