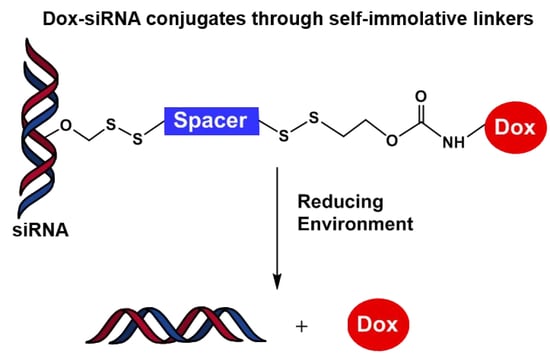
Conjugation of Doxorubicin to siRNA Through Disulfide-based Self-immolative Linkers
Abstract
:1. Introduction
2. Results and Discussion
2.1. Synthesis of Dox-RNA Conjugates
2.1.1. Synthesis of Alkylbis(disulfanylpyridine) Reagents 1 and 2
2.1.2. Synthesis of 2′-O-activated RNAs
2.1.3. Conjugation of Doxorubicin to 2′-O-PySSR1SSMe and 2′-O-PySSR2SSMe RNAs
2.2. Biophysical Properties of Dox-siRNA Conjugates
2.2.1. Thermal Denaturation Studies of Dox-siRNAs
2.2.2. Circular Dichroism Studies of Dox-siRNAs
2.3. Stability of Dox-siRNAs in Human Serum
2.4. Unmasking of a Dox-RNA Conjugate under Reductive Environment
3. Materials and Methods
3.1. General Methods
3.2. 1,5-bis(pyridin-2-yldisulfanyl)pentane 1
3.3. Bis(pyridin-2-yldisulfanyl)-p-xylene 2
3.4. Solid-phase Synthesis of RNA Oligonucleotides
3.5. Synthesis of Modified 2′-O- PySSRSSMe RNAs G1-R1, C11-R1, G1-R2, C11-R2 and G1C11-R2
3.6. Synthesis of Dox-RNA Conjugates G1-R1-Dox, C11-R1-Dox, G1-R2-Dox, C11-R2-Dox and G1C11-R2-Dox
3.7. Thermal Denaturation Studies of Dox-siRNA Duplexes
3.8. CD Spectroscopy Studies of Dox-siRNA Duplexes
3.9. Serum Stability of Dox-siRNA Duplexes
3.10. Unmasking of G1-R2-Dox Conjugate in the Presence of TCEP
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Gottesman, M.M.; Fojo, T.; Bates, S.E. Multidrug resistance in cancer: Role of ATP-dependent transporters. Nat. Rev. Cancer. 2002, 2, 48–58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsouris, V.; Joo, M.K.; Kim, S.H.; Kwon, I.C.; Won, Y.Y. Nano carriers that enable co-delivery of chemotherapy and RNAi agents for treatment of drug-resistant cancers. Biotechnol. Adv. 2014, 32, 1037–1050. [Google Scholar] [CrossRef] [PubMed]
- Saraswathy, M.; Gong, S. Recent developments in the co-delivery of siRNA and small molecule anticancer drugs for cancer treatment. Mater. Today 2014, 17, 298–306. [Google Scholar] [CrossRef]
- Minotti, G.; Menna, P.; Salvatorelli, E.; Cairo, G.; Gianni, L. Anthracyclines: Molecular advances and pharmacologic developments in antitumor activity and cardiotoxicity. Pharmacol. Rev. 2004, 56, 185–229. [Google Scholar] [CrossRef] [Green Version]
- Carvalho, C.; Santos, R.X.; Cardoso, S.; Correia, S.; Oliveira, P.J.; Santos, M.S.; Moreira, P.I. Doxorubicin: The good, the bad and the ugly effect. Curr. Med. Chem. 2009, 16, 3267–3285. [Google Scholar] [CrossRef]
- Mitry, M.A.; Edwards, J.G. Doxorubicin induced heart failure: Phenotype and molecular mechanisms. Int. J. Cardiol. Heart Vasc. 2016, 10, 17–24. [Google Scholar] [CrossRef] [Green Version]
- Haussecker, D.; Kay, M.A. RNA interference. Drugging RNAi. Science 2015, 347, 1069–1070. [Google Scholar] [CrossRef]
- Wittrup, A.; Lieberman, J. Knocking down disease: A progress report on siRNA therapeutics. Nat. Rev. Genet. 2015, 16, 543–552. [Google Scholar] [CrossRef]
- Li, J.; Wang, Y.; Zhu, Y.; Oupicky, D. Recent advances in delivery of drug-nucleic acid combinations for cancer treatment. J. Control. Release 2013, 172, 589–600. [Google Scholar] [CrossRef] [Green Version]
- Ming, X.; Laing, B. Bioconjugates for targeted delivery of therapeutic oligonucleotides. Adv. Drug Deliv. Rev. 2015, 87, 81–89. [Google Scholar] [CrossRef] [Green Version]
- Butt, A.M.; Amin, M.C.I.M.; Katas, H.; Abdul Murad, N.A.; Jamal, R.; Kesharwani, P. Doxorubicin and siRNA Codelivery via Chitosan-Coated pH-Responsive Mixed Micellar Polyplexes for Enhanced Cancer Therapy in Multidrug-Resistant Tumors. Mol. Pharm. 2016, 13, 4179–4190. [Google Scholar] [CrossRef] [PubMed]
- Babu, A.; Munshi, A.; Ramesh, R. Combinatorial therapeutic approaches with RNAi and anticancer drugs using nanodrug delivery systems. Drug Dev. Ind. Pharm. 2017, 43, 1391–1401. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Feng, S.S. Nanocarriers for delivery of siRNA and co-delivery of siRNA and other therapeutic agents. Nanomed. J. 2015, 10, 2199–2228. [Google Scholar] [CrossRef]
- Xiao, B.; Ma, L.; Merlin, D. Nanoparticle-mediated co-delivery of chemotherapeutic agent and siRNA for combination cancer therapy. Expert Opin. Drug Deliv. 2017, 14, 65–73. [Google Scholar] [CrossRef]
- Wu, M.; Li, J.; Lin, X.; Wei, Z.; Zhang, D.; Zhao, B.; Liu, X.; Liu, J. Reduction/photo dual-responsive polymeric prodrug nanoparticles for programmed siRNA and doxorubicin delivery. Biomater. Sci. 2018, 6, 1457–1468. [Google Scholar] [CrossRef]
- Alinejad, V.; Hossein Somi, M.; Baradaran, B.; Akbarzadeh, P.; Atyabi, F.; Kazerooni, H.; Samadi Kafil, H.; Aghebati Maleki, L.; Siah Mansouri, H.; Yousefi, M. Co-delivery of IL17RB siRNA and doxorubicin by chitosan-based nanoparticles for enhanced anticancer efficacy in breast cancer cells. Biomed. Pharmacother. 2016, 83, 229–240. [Google Scholar] [CrossRef]
- Ma, X.; Teh, C.; Zhang, Q.; Borah, P.; Choong, C.; Korzh, V.; Zhao, Y. Redox-Responsive Mesoporous Silica Nanoparticles: A Physiologically Sensitive Codelivery Vehicle for siRNA and Doxorubicin. Antioxid. Redox Signal. 2013, 21, 707–722. [Google Scholar] [CrossRef]
- Meng, H.; Mai, W.X.; Zhang, H.Y.; Xue, M.; Xia, T.; Lin, S.J.; Wang, X.; Zhao, Y.; Ji, Z.X.; Zink, J.I.; et al. Codelivery of an Optimal Drug/siRNA Combination Using Mesoporous Silica Nanoparticles To Overcome Drug Resistance in Breast Cancer in Vitro and in Vivo. ACS Nano 2013, 7, 994–1005. [Google Scholar] [CrossRef] [Green Version]
- Zhou, X.; Chen, L.; Nie, W.; Wang, W.; Qin, M.; Mo, X.; Wang, H.; He, C. Dual-Responsive Mesoporous Silica Nanoparticles Mediated Codelivery of Doxorubicin and Bcl-2 SiRNA for Targeted Treatment of Breast Cancer. J. Phys. Chem. C 2016, 120, 22375–22387. [Google Scholar] [CrossRef]
- Xia, Y.; Xu, T.; Wang, C.; Li, Y.; Lin, Z.; Zhao, M.; Zhu, B. Novel functionalized nanoparticles for tumor-targeting co-delivery of doxorubicin and siRNA to enhance cancer therapy. Int. J. Nanomed. 2018, 13, 143–159. [Google Scholar] [CrossRef] [Green Version]
- Zang, C.; Wang, H.; Li, T.; Zhang, Y.; Li, J.; Shang, M.; Du, J.; Xi, Z.; Zhou, C. A light-responsive, self-immolative linker for controlled drug delivery via peptide- and protein-drug conjugates. Chem. Sci. 2019, 10, 8973–8980. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alouane, A.; Labruere, R.; Le Saux, T.; Schmidt, F.; Jullien, L. Self-immolative spacers: Kinetic aspects, structure-property relationships, and applications. Angew. Chem. Int. Ed. Engl. 2015, 54, 7492–7509. [Google Scholar] [CrossRef] [PubMed]
- Deng, Z.; Hu, J.; Liu, S. Disulfide-Based Self-Immolative Linkers and Functional Bioconjugates for Biological Applications. Macromol. Rapid Commun. 2020, 41, 1900531. [Google Scholar] [CrossRef] [Green Version]
- Jain, A.K.; Gund, M.G.; Desai, D.C.; Borhade, N.; Senthilkumar, S.P.; Dhiman, M.; Mangu, N.K.; Mali, S.V.; Dubash, N.P.; Halder, S.; et al. Mutual prodrugs containing bio-cleavable and drug releasable disulfide linkers. Bioorg. Chem. 2013, 49, 40–48. [Google Scholar] [CrossRef] [PubMed]
- Gund, M.; Khanna, A.; Dubash, N.; Damre, A.; Singh, K.S.; Satyam, A. Water-soluble prodrugs of paclitaxel containing self-immolative disulfide linkers. Bioorg. Med. Chem. Lett. 2015, 25, 122–127. [Google Scholar] [CrossRef]
- Perry, R.R.; Mazetta, J.; Levin, M.; Barranco, S.C. Glutathione levels and variability in breast tumors and normal tissue. Cancer 1993, 72, 783–787. [Google Scholar] [CrossRef]
- Stasińska, A.R.; Putaj, P.; Chmielewski, M.K. Disulfide bridge as a linker in nucleic acids’ bioconjugation. Part I: An overview of synthetic strategies. Bioorg. Chem. 2019, 92, 103223. [Google Scholar] [CrossRef]
- Stasińska, A.R.; Putaj, P.; Chmielewski, M.K. Disulfide bridge as a linker in nucleic acids’ bioconjugation. Part II: A summary of practical applications. Bioorg. Chem. 2020, 95, 103518. [Google Scholar] [CrossRef]
- Ramon, A.L.; Bertrand, J.R.; de Martimprey, H.; Bernard, G.; Ponchel, G.; Malvy, C.; Vauthier, C. siRNA associated with immunonanoparticles directed against cd99 antigen improves gene expression inhibition in vivo in Ewing’s sarcoma. J. Mol. Recognit. 2013, 26, 318–329. [Google Scholar] [CrossRef]
- Gauthier, F.; Malher, A.; Vasseur, J.J.; Dupouy, C.; Debart, F. Conjugation of small molecules to RNA using a reducible disulfide linker attached at 2′OH position via a carbamate function. Eur. J. Org. Chem. 2019, 5636–5645. [Google Scholar] [CrossRef]
- Gauthier, F.; Claveau, S.; Bertrand, J.-R.; Vasseur, J.-J.; Dupouy, C.; Debart, F. Gymnotic delivery and gene silencing activity of reduction-responsive siRNAs bearing lipophilic disulfide-containing modifications at 2′-position. Bioorg. Med. Chem. 2018, 26, 4635–4643. [Google Scholar] [CrossRef] [PubMed]
- Biscans, A.; Rouanet, S.; Vasseur, J.J.; Dupouy, C.; Debart, F. A versatile post-synthetic method on a solid support for the synthesis of RNA containing reduction-responsive modifications. Org. Biomol. Chem. 2016, 14, 7010–7017. [Google Scholar] [CrossRef] [PubMed]
- Satyam, A. Design and synthesis of releasable folate–drug conjugates using a novel heterobifunctional disulfide-containing linker. Bioorg. Med. Chem. Lett. 2008, 18, 3196–3199. [Google Scholar] [CrossRef] [PubMed]
- Jones, L.R.; Goun, E.A.; Shinde, R.; Rothbard, J.B.; Contag, C.H.; Wender, P.A. Releasable Luciferin−Transporter Conjugates: Tools for the Real-Time Analysis of Cellular Uptake and Release. J. Am. Chem. Soc. 2006, 128, 6526–6527. [Google Scholar] [CrossRef]
- Batisse, C.; Dransart, E.; Ait Sarkouh, R.; Brulle, L.; Bai, S.-K.; Godefroy, S.; Johannes, L.; Schmidt, F. A new delivery system for auristatin in STxB-drug conjugate therapy. Eur. J. Med. Chem. 2015, 95, 483–491. [Google Scholar] [CrossRef] [PubMed]
- Lapeyre, M.; Leprince, J.; Massonneau, M.; Oulyadi, H.; Renard, P.-Y.; Romieu, A.; Turcatti, G.; Vaudry, H. Aryldithioethyloxycarbonyl (Ardec): A New Family of Amine Protecting Groups Removable under Mild Reducing Conditions and Their Applications to Peptide Synthesis. Chem. Eur. J. 2006, 12, 3655–3671. [Google Scholar] [CrossRef] [PubMed]
- Latorre, A.; Couleaud, P.; Aires, A.; Cortajarena, A.L.; Somoza, A. Multifunctionalization of magnetic nanoparticles for controlled drug release: A general approach. Eur. J. Med. Chem. 2014, 82, 355–362. [Google Scholar] [CrossRef]
- Riggi, N.; Cironi, L.; Provero, P.; Suva, M.L.; Kaloulis, K.; Garcia-Echeverria, C.; Hoffmann, F.; Trumpp, A.; Stamenkovic, I. Development of Ewing’s sarcoma from primary bone marrow-derived mesenchymal progenitor cells. Cancer Res. 2005, 65, 11459–11468. [Google Scholar] [CrossRef] [Green Version]
- Lavergne, T.; Bertrand, J.-R.; Vasseur, J.-J.; Debart, F. A base-labile group for 2′-OH protection of ribonucleosides: A major challenge for RNA synthesis. Chem. Eur. J. 2008, 14, 9135–9138. [Google Scholar] [CrossRef]
- Gauthier, F.; Beltran, F.; Biscans, A.; Debart, F.; Dupouy, C.; Vasseur, J.-J. A 2′,2′-disulfide-bridged dinucleotide conformationally locks RNA hairpins. Org. Biomol. Chem. 2018, 16, 3181–3188. [Google Scholar] [CrossRef]
- Tian, Y.; Bromberg, L.; Lin, S.N.; Hatton, T.A.; Tam, K.C. Complexation and release of doxorubicin from its complexes with pluronic P85-b-poly(acrylic acid) block copolymers. J. Control. Release 2007, 121, 137–145. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are not available from the authors. |










| RNA | Sequence 5′ → 3′ 1 | MALDI-TOF MS 2 | Conjugation Yield (%) 3 | |
|---|---|---|---|---|
| Calcd. | Found | |||
| S | GCA GCA GAA CCC UUC UUA UGA | / | / | / |
| G1-R1 | GR1CA GCA GAA CCC UUC UUA UGA | 6945.6 | 6944.4 | 32 |
| C11-R1 | GCA GCA GAA CCR1UUC UUA UGA | 6945.6 | 6944.1 | 28 |
| G1-R2 | GR2CA GCA GAA CCC UUC UUA UGA | 6979.6 | 6978.1 | 24 |
| C11-R2 | GCA GCA GAA CCR2C UUC UUA UGA | 6979.6 | 6978.6 | 35 |
| G1C11-R2 | GR2CA GCA GAA CCR2C UUC UUA UGA | 7296.2 | 7294.3 | 15 |
| RNA | Sequence 5′ → 3′ 1 | MALDI-TOF MS 2 | Conjugation Yield (%) 3 | Recovery Yield (%) 4 | Tm5 (°C) | Half-life HS (min) 6 | |
|---|---|---|---|---|---|---|---|
| Calcd. | Found | ||||||
| S G1-R1-Dox | GCAGCAGAACCCUUCUUAUGA GDoxCAGCAGAACCCUUCUUAUGA | / 7482.1 | / 7482.8 | / 91 | / 39 | 75.6 70.1 | <5.0 13 |
| C11-R1-Dox | GCAGCAGAACCDoxCUUCUUAUGA | 7482.1 | 7484.0 | 80 | 46 | 70.5 | 38 |
| G1-R2-Dox | GDoxCAGCAGAACCCUUCUUAUGA | 7517.1 | 7516.5 | 80 | 49 | 73.4 | 20 |
| C11-R2-Dox | GCAGCAGAACCDoxCUUCUUAUGA | 7517.1 | 7515.3 | 69 | 43 | 72.9 | 10 |
| G1C11-R2-Dox | GDoxCAGCAGAACCDoxCUUCUUAUGA | 8375.1 | 8375.3 | 62 | 26 | 67.8 | 48 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gauthier, F.; Bertrand, J.-R.; Vasseur, J.-J.; Dupouy, C.; Debart, F. Conjugation of Doxorubicin to siRNA Through Disulfide-based Self-immolative Linkers. Molecules 2020, 25, 2714. https://doi.org/10.3390/molecules25112714
Gauthier F, Bertrand J-R, Vasseur J-J, Dupouy C, Debart F. Conjugation of Doxorubicin to siRNA Through Disulfide-based Self-immolative Linkers. Molecules. 2020; 25(11):2714. https://doi.org/10.3390/molecules25112714
Chicago/Turabian StyleGauthier, Florian, Jean-Rémi Bertrand, Jean-Jacques Vasseur, Christelle Dupouy, and Françoise Debart. 2020. "Conjugation of Doxorubicin to siRNA Through Disulfide-based Self-immolative Linkers" Molecules 25, no. 11: 2714. https://doi.org/10.3390/molecules25112714
APA StyleGauthier, F., Bertrand, J.-R., Vasseur, J.-J., Dupouy, C., & Debart, F. (2020). Conjugation of Doxorubicin to siRNA Through Disulfide-based Self-immolative Linkers. Molecules, 25(11), 2714. https://doi.org/10.3390/molecules25112714