Chemical Composition, Antimicrobial Properties of Siparuna guianensis Essential Oil and a Molecular Docking and Dynamics Molecular Study of its Major Chemical Constituent
Abstract
:1. Introduction
2. Results and Discussion
2.1. Yield and Chemical Composition
2.2. Antimicrobial Activity
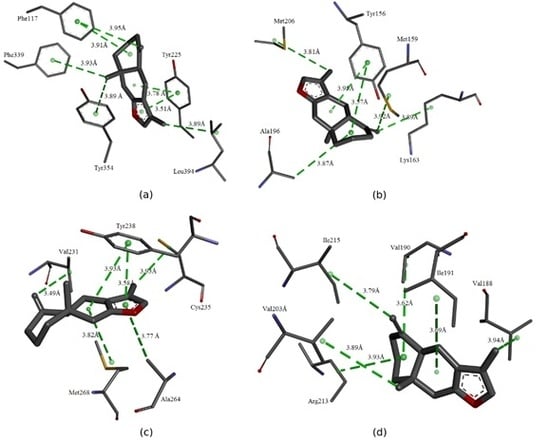
3. Interaction Mechanism
3.1. Molecular Binding Mode
3.2. Analysis of Complexes Stability
3.3. Free Energy Calculations Using MM/GBSA Approach
4. Materials and Methods
4.1. Preparation and Characterization of the Siparuna guianensis Sample
4.1.1. Botanical Information of the Sample
4.1.2. Extraction Procedure: Hydrodistillation
4.2. Analysis of Volatile Compounds
4.3. Analysis of In vitro Antimicrobial Activity
4.4. Evaluation of the Sample Sensitivity by the Disk Diffusion Method
4.5. Determination of Minimum Inhibitory Concentration (MIC)
4.6. Statistical Analysis
5. Molecular Docking and Dynamics Molecular Simulations
5.1. Molecular Docking
5.2. Molecular Dynamics (MD) Simulation
5.3. Free Energy Calculations
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- O’Brien, C.R.; Malik, R.; Globan, M.; Reppas, G.; McCowan, C.; Fyfe, J.A. Feline leprosy due to Mycobacterium lepraemurium: Further clinical and molecular characterisation of 23 previously reported cases and an additional 42 cases. J. Feline Med. Surg. 2017, 19, 737–746. [Google Scholar] [CrossRef] [PubMed]
- Hmama, Z.; Peña-Díaz, S.; Joseph, S.; Av-Gay, Y. Immunoevasion and immunosuppression of the macrophage by Mycobacterium tuberculosis. Immunol. Rev. 2015, 264, 220–232. [Google Scholar] [CrossRef]
- Schofield, D.A.; Wray, D.J.; Molineux, I.J. Isolation and development of bioluminescent reporter phages for bacterial dysentery. Eur. J. Clin. Microbiol. Infect. Dis. 2014, 34, 395–403. [Google Scholar] [CrossRef]
- Fairley, C.K.; Hocking, J.S.; Zhang, L.; Chow, E.P.F. Frequent transmission of gonorrhea in men who have sex with men. Emerg. Infect. Dis. 2017, 23, 102–104. [Google Scholar] [CrossRef] [PubMed]
- Murray, B.E. The life and times of the Enterococcus. Clin. Microbiol. Rev. 1990, 3, 46–65. [Google Scholar] [CrossRef]
- Fasugba, O.; Gardner, A.; Mitchell, B.G.; Mnatzaganian, G. Ciprofloxacin resistance in community- and hospital-acquired Escherichia coli urinary tract infections: A systematic review and meta-analysis of observational studies. BMC Infect. Dis. 2015, 15. [Google Scholar] [CrossRef] [Green Version]
- Shemer, A.; Gupta, A.K.; Amichai, B.; Baum, S.; Barzilai, A.; Farhi, R.; Kaplan, Y.; MacLeod, M.A. Increased Risk of Tinea Pedis and Onychomycosis Among Swimming Pool Employees in Netanya Area, Israel. Mycopathologia 2016, 181, 851–856. [Google Scholar] [CrossRef] [PubMed]
- Marty, F.M.; Ostrosky-Zeichner, L.; Cornely, O.A.; Mullane, K.M.; Perfect, J.R.; Thompson, G.R.; Alangaden, G.J.; Brown, J.M.; Fredricks, D.N.; Heinz, W.J.; et al. Isavuconazole treatment for mucormycosis: A single-arm open-label trial and case-control analysis. Lancet Infect. Dis. 2016, 16, 828–837. [Google Scholar] [CrossRef]
- Kullberg, B.J.; Arendrup, M.C. Invasive Candidiasis. N. Engl. J. Med. 2015, 373, 1445–1456. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Falsetta, M.L.; Klein, M.I.; Colonne, P.M.; Scott-Anne, K.; Gregoire, S.; Pai, C.H.; Gonzalez-Begne, M.; Watson, G.; Krysan, D.J.; Bowen, W.H.; et al. Symbiotic relationship between Streptococcus mutans and Candida albicans synergizes virulence of plaque biofilms in vivo. Infect. Immun. 2014, 82, 1968–1981. [Google Scholar] [CrossRef] [Green Version]
- Kaye, K.S.; Pogue, J.M. Infections Caused by Resistant Gram-Negative Bacteria: Epidemiology and Management. Pharmacotherapy 2015, 35, 949–962. [Google Scholar] [CrossRef]
- Łysakowska, M.E.; Sienkiewicz, M.; Banaszek, K.; Sokołowski, J.; McPhee, D.J. The sensitivity of endodontic enterococcus Spp. Strains to geranium essential oil. Molecules 2015, 20, 22881–22889. [Google Scholar] [CrossRef] [PubMed]
- Marrufo, T.; Nazzaro, F.; Mancini, E.; Fratianni, F.; Coppola, R.; De Martino, L.; Agostinho, A.B.; De Feo, V. Chemical composition and biological activity of the essential oil from leaves of Moringa oleifera Lam. cultivated in Mozambique. Molecules 2013, 18, 10989–11000. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mandal, S.; Mandal, M. Coriander (Coriandrum sativum L.) essential oil: Chemistry and biological activity. Asian Pac. J. Trop. Biomed. 2015, 5, 421–428. [Google Scholar] [CrossRef] [Green Version]
- Lai, P.; Rao, H.; Gao, Y. Chemical Composition, Cytotoxic, Antimicrobial and Antioxidant Activities of Essential oil from Anthriscus caucalis M. Bieb Grown in China. Rec. Nat. Prod. 2018, 3, 290–294. [Google Scholar] [CrossRef]
- Bayala, B.; Bassole, I.H.N.; Gnoula, C.; Nebie, R.; Yonli, A.; Morel, L.; Figueredo, G.; Nikiema, J.B.; Lobaccaro, J.M.A.; Simpore, J. Chemical composition, antioxidant, anti-inflammatory and anti-proliferative activities of essential oils of plants from Burkina Faso. PLoS One 2014, 9, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Arranz, E.; Jaime, L.; López de las Hazas, M.C.; Reglero, G.; Santoyo, S. Supercritical fluid extraction as an alternative process to obtain essential oils with anti-inflammatory properties from marjoram and sweet basil. Ind. Crops Prod. 2015, 67, 121–129. [Google Scholar] [CrossRef]
- Oliveira, M.S.d.; Almeida, M.M.; Salazar, M.d.L.A.R.; Pires, F.C.S.; Bezerra, F.W.F.; Cunha, V.M.B.; Cordeiro, R.M.; Urbina, G.R.O.; Silva, M.P.d.; Silva, A.P.S.e. Potential of Medicinal Use of Essential Oils from Aromatic Plants. In Potential of Essential Oils; Hany, A.E.-S., Ed.; InTech: Londo, UK, 2018; p. 150. [Google Scholar]
- Zhang, H.-Y.; Gao, Y.; Lai, P.-X. Chemical Composition, Antioxidant, Antimicrobial and Cytotoxic Activities of Essential Oil from Premna microphylla Turczaninow. Molecules 2017, 22, 381. [Google Scholar] [CrossRef] [Green Version]
- Mothana, R.A.; Noman, O.M.; Al-Sheddi, E.S.; Khaled, J.M.; Al-Said, M.S.; Al-Rehaily, A.J. Chemical composition, in vitro antimicrobial, free-radical-scavenging and antioxidant activities of the essential oil of Leucas inflata Benth. Molecules 2017, 22, 367. [Google Scholar] [CrossRef] [Green Version]
- Diao, W.-R.; Hu, Q.-P.; Zhang, H.; Xu, J.-G. Chemical composition, antibacterial activity and mechanism of action of essential oil from seeds of fennel (Foeniculum vulgare Mill.). Food Control 2014, 35, 109–116. [Google Scholar] [CrossRef]
- Pesavento, G.; Calonico, C.; Bilia, A.R.; Barnabei, M.; Calesini, F.; Addona, R.; Mencarelli, L.; Carmagnini, L.; Di Martino, M.C.; Lo Nostro, A. Antibacterial activity of Oregano, Rosmarinus and Thymus essential oils against Staphylococcus aureus and Listeria monocytogenes in beef meatballs. Food Control 2015, 54, 188–199. [Google Scholar] [CrossRef]
- Da Silva Bomfim, N.; Nakassugi, L.P.; Faggion Pinheiro Oliveira, J.; Kohiyama, C.Y.; Mossini, S.A.G.; Grespan, R.; Nerilo, S.B.; Mallmann, C.A.; Alves Abreu Filho, B.; Machinski, M. Antifungal activity and inhibition of fumonisin production by Rosmarinus officinalis L. essential oil in Fusarium verticillioides (Sacc.) Nirenberg. Food Chem. 2015, 166, 330–336. [Google Scholar] [CrossRef] [Green Version]
- Freires, I.D.A.; Murata, R.M.; Furletti, V.F.; Sartoratto, A.; De Alencar, S.M.; Figueira, G.M.; Rodrigues, J.A.D.O.; Duarte, M.C.T.; Rosalen, P.L. Coriandrum sativum L. (Coriander) essential oil: Antifungal activity and mode of action on Candida spp., and molecular targets affected in human whole-genome expression. PLoS ONE 2014, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bektaş, E.; Daferera, D.; Sökmen, M.; Serdar, G.; Ertürk, M.; Polissiou, M.G.; Sökmen, A. In vitro antimicrobial, antioxidant, and antiviral activities of the essential oil and various extracts from thymus nummularis M. Bieb. Indian J. Tradit. Knowl. 2016, 15, 403–410. [Google Scholar]
- Pourghanbari, G.; Nili, H.; Moattari, A.; Mohammadi, A.; Iraji, A. Antiviral activity of the oseltamivir and Melissa officinalis L. essential oil against avian influenza A virus (H9N2). VirusDisease 2016, 27, 170–178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Masoumi-Ardakani, Y.; Mandegary, A.; Esmaeilpour, K.; Najafipour, H.; Sharififar, F.; Pakravanan, M.; Ghazvini, H. Chemical composition, anticonvulsant activity, and toxicity of essential oil and methanolic extract of Elettaria cardamomum. Planta Med. 2016, 82, 1482–1486. [Google Scholar] [CrossRef]
- Randrianarivo, E.; Maggi, F.; Nicoletti, M.; Rasoanaivo, P. Evaluation of the anticonvulsant activity of the essential oil of Myrothamnus moschatus in convulsion induced by pentylenetetrazole and picrotoxin. Asian Pac. J. Trop. Biomed. 2016, 6, 501–505. [Google Scholar] [CrossRef] [Green Version]
- Mendes, S.S.; Bomfim, R.R.; Jesus, H.C.R.; Alves, P.B.; Blank, A.F.; Estevam, C.S.; Antoniolli, A.R.; Thomazzi, S.M. Evaluation of the analgesic and anti-inflammatory effects of the essential oil of Lippia gracilis leaves. J. Ethnopharmacol. 2010, 129, 391–397. [Google Scholar] [CrossRef]
- Rodriguez, O.E.; Sánchez, R.M.; Verde, M.J.; Núñez, M.A.; Castro, R.; Chávez, A. Obtaining of the essential oil of Syzygium aromaticum, identification of eugenol and its effect on Streptococcus mutans. J. Oral Res. 2014, 3, 218–224. [Google Scholar] [CrossRef] [Green Version]
- Zeng, Z.; Zhang, S.; Wang, H.; Piao, X. Essential oil and aromatic plants as feed additives in non-ruminant nutrition: A review. J. Anim. Sci. Biotechnol. 2015, 6. [Google Scholar] [CrossRef] [Green Version]
- Salgado, A.P.S.P.; Cardoso, M.D.G.; De Souza, P.E.; De Souza, J.A.; Abreu, C.M.P.; Pinto, J.E.B.P. De Folhas De Eucalyptus SOBRE Fusarium oxysporum, Fungitoxic Activity Evaluation of Essential Leaf Oils of Eucalyptus on Fusarium oxysporum, Botrytis cinerea and Bipolaris sorokiniana. Ciênc. Agrotec. 2003, 27, 249–254. [Google Scholar] [CrossRef]
- Renner, S.S.; Hausner, G. Siparunaceae, 1st ed.; New York Botanical Garden: New York, NY, USA, 2005. [Google Scholar]
- Valentini, C.M.; Rodríguez-Ortíz, C.; Coelho, M.F. Siparuna guianensis Aublet (negramina): Uma revisão. Rev. Bras. Plantas Med. 2010, 12, 96–104. [Google Scholar] [CrossRef] [Green Version]
- Melo, D.; Miranda, M.; Junior, W.; Alcoba, A.; Andrade, P.; Silva, T.D.S.; Cazal, C.; Martins, C. Anticariogenic and Antimycobacterial Activities of the Essential Oil of Siparuna guianensis Aublet (Siparunaceae). Orbital - Electron. J. Chem. 2017, 9. [Google Scholar] [CrossRef]
- Andrade, M.A.; Cardoso, M.D.G.; Gomes, M.d.S.; Azeredo, C.M.O.d.; Batista, L.R.; Soares, M.J.; Rodrigues, L.M.A.; Figueiredo, A.C.S. Biological activity of the essential oils from Cinnamodendron dinisii and Siparuna guianensis. Brazilian J. Microbiol. 2015, 46, 189–194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Souza Moura, W.; de Souza, S.R.; Campos, F.S.; Sander Rodrigues Cangussu, A.; Macedo Sobrinho Santos, E.; Silva Andrade, B.; Borges Gomes, C.H.; Fernandes Viana, K.; Haddi, K.; Oliveira, E.E.; et al. Antibacterial activity of Siparuna guianensis essential oil mediated by impairment of membrane permeability and replication of pathogenic bacteria. Ind. Crops Prod. 2020, 146, 112142. [Google Scholar] [CrossRef]
- Issn, O. Revista de Biología Tropical. Rev. Biol. Trop. 2000, 11, 1–15. [Google Scholar]
- de Bessa, N.G.F.; Pereira, M.E.A.B.; Ferraz, V.; Poletto, K.Q.; Junior, A.I.F.C.; Alves, A. Antimicrobial activity and medicinal biomass of Siparuna guianensis in Brazilian Cerrado forest, a global hotspot. J. Med. Plants Res. 2015, 9, 968–980. [Google Scholar] [CrossRef]
- Andrade, M.; das Graças Cardoso, M.; de Andrade, J.; Silva, L.; Teixeira, M.; Valério Resende, J.; da Silva Figueiredo, A.; Barroso, J. Chemical Composition and Antioxidant Activity of Essential Oils from Cinnamodendron dinisii Schwacke and Siparuna guianensis Aublet. Antioxidants 2013, 2, 384–397. [Google Scholar] [CrossRef]
- Fischer, D.C.H.; Limberger, R.P.; Henriques, A.T.; Moreno, P.R.H. Essential oils from fruits and Leaves of Siparuna guianensis (Aubl.) Tulasne from Southeastern Brazil. J. Essent. Oil Res. 2005, 17, 101–102. [Google Scholar] [CrossRef]
- Zoghbi, M.d.G.B.; Andrade, E.H.A.; Santos, A.S.; Silva, M.H.L.; Maia, J.G.S. Essential Oils of Siparuna guianensis Aubl. J. Essent. Oil Res. 1998, 10, 543–546. [Google Scholar] [CrossRef]
- Machado, S.M.F.; Ribeiro, V.A.F.A.; Militão, J.S.L.T.; De Morais, S.M.; Machado, M.I.L. Seasonal variation of (E)-nerolidol in siparuna guianensis aublet and13C-NMR spectral assignments of (E)- and (Z)-nerolidol. J. Essent. Oil Res. 1998, 10, 708–710. [Google Scholar] [CrossRef]
- Viana, F.A.; Andrade-Neto, M.; Pouliquen, Y.B.M.; Uchoa, D.E.A.; Sobral, M.M.S.Z.; de Morais, S.M. Essential oil of siparuna guianensis aublet from the amazon region of Brazil. J. Essent. Oil Res. 2002, 14, 60–62. [Google Scholar] [CrossRef]
- Andrade, E.H.A.; Zoghbi, M.G.B.; Machado, L.B. Seasonal variation of germacrone and atractylone in the essential oil of Siparuna guianensis Aublet. Rev. Bras. Plantas Med. 2004, 6, 62–64. [Google Scholar]
- Adams, R.P. Identification of Essential Oil Components by Gas Chromatography/Mass Spectroscopy, 4th ed.; Adams, R.P., Ed.; Allured Publishing Corporation: Carol Stream, IL, USA, 28 February 2007; ISBN 1932633219. [Google Scholar]
- Stein, S.; Mirokhin, D.; Tchekhovskoi, D.; Mallard, G.; Mikaia, A.; Zaikin, V.; Sparkmanm, D. The NIST Mass Spectral Search Program for the Nist/Epa/Nih Mass Spectra Library; Standard Reference Data Program of the National Institute of Standards and Technology: Gaithers-burg, MD, USA, 2011. [Google Scholar]
- Smith-Palmer, A.; Stewart, J.; Fyfe, L. Antimicrobial properties of plant essential oils and essences against five important food-borne pathogens. Lett. Appl. Microbiol. 1998, 26, 118–122. [Google Scholar] [CrossRef]
- Cavaleiro, C.; Salgueiro, L.; Gonçalves, M.J.; Hrimpeng, K.; Pinto, J.; Pinto, E. Antifungal activity of the essential oil of Angelica major against Candida, Cryptococcus, Aspergillus and dermatophyte species. J. Nat. Med. 2015, 69, 241–248. [Google Scholar] [CrossRef]
- Resch, M.; Steigel, A.; Chen, Z.L.; Bauer, R. 5-lipoxygenase and cyclooxygenase-1 inhibitory active compounds from Atractylodes lancea. J. Nat. Prod. 1998, 61, 347–350. [Google Scholar] [CrossRef]
- Ogunwande, I.A.; Olawore, N.O.; Ekundayo, O.; Walker, T.M.; Schmidt, J.M.; Setzer, W.N. Studies on the essential oils composition, antibacterial and cytotoxicity of Eugenia uniflora L. Int. J. Aromather. 2005, 15, 147–152. [Google Scholar] [CrossRef]
- Govindarajan, M.; Rajeswary, M.; Senthilmurugan, S.; Vijayan, P.; Alharbi, N.S.; Kadaikunnan, S.; Khaled, J.M.; Benelli, G. Curzerene, trans-β-elemenone, and γ-elemene as effective larvicides against Anopheles subpictus, Aedes albopictus, and Culex tritaeniorhynchus: Toxicity on non-target aquatic predators. Environ. Sci. Pollut. Res. 2018, 25, 10272–10282. [Google Scholar] [CrossRef] [PubMed]
- Gurgel, E.S.C.; de Oliveira, M.S.; Souza, M.C.; Silva, S.G.d.; de Mendonça, M.S.; Souza Filho, A.P.d.S. Chemical compositions and herbicidal (phytotoxic) activity of essential oils of three Copaifera species (Leguminosae-Caesalpinoideae) from Amazon-Brazil. Ind. Crops Prod. 2019, 142, 111850. [Google Scholar] [CrossRef]
- Ferreira, O.O.; da Cruz, J.N.; Franco, C.d.J.P.; Silva, S.G.; da Costa, W.A.; de Oliveira, M.S.; Andrade, E.H.d.A. First Report on Yield and Chemical Composition of Essential Oil Extracted from Myrcia eximia DC (Myrtaceae) from the Brazilian Amazon. Molecules 2020, 25, 783. [Google Scholar] [CrossRef] [Green Version]
- Clinical and Laboratory Standards Institute (CLSI). Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically; Approved Standard—Seventh Edition; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2006; Volume 2, ISBN 1-56238-587-9. [Google Scholar]
- Knezevic, P.; Aleksic, V.; Simin, N.; Svircev, E.; Petrovic, A.; Mimica-Dukic, N. Antimicrobial activity of Eucalyptus camaldulensis essential oils and their interactions with conventional antimicrobial agents against multi-drug resistant Acinetobacter baumannii. J. Ethnopharmacol. 2016, 178, 125–136. [Google Scholar] [CrossRef]
- Sogabe, S.; Masubuchi, M.; Sakata, K.; Fukami, T.A.; Morikami, K.; Shiratori, Y.; Ebiike, H.; Kawasaki, K.; Aoki, Y.; Shimma, N.; et al. Crystal Structures of Candida albicans N-Myristoyltransferase with Two Distinct Inhibitors. Chem. Biol. 2002, 9, 1119–1128. [Google Scholar] [CrossRef] [Green Version]
- Qiu, X.; Abdel-Meguid, S.S.; Janson, C.A.; Court, R.I.; Smyth, M.G.; Payne, D.J. Molecular basis for triclosan activity involves a flipping loop in the active site. Protein Sci. 2008, 8, 2529–2532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramón-Maiques, S.; Marina, A.; Guinot, A.; Gil-Ortiz, F.; Uriarte, M.; Fita, I.; Rubio, V. Substrate binding and catalysis in carbamate kinase ascertained by crystallographic and site-directed mutagenesis studies: Movements and significance of a unique globular subdomain of this key enzyme for fermentative ATP production in bacteria. J. Mol. Biol. 2010, 397, 1261–1275. [Google Scholar] [CrossRef] [PubMed]
- Wallock-Richards, D.J.; Marles-Wright, J.; Clarke, D.J.; Maitra, A.; Dodds, M.; Hanley, B.; Campopiano, D.J. Molecular basis of Streptococcus mutans sortase A inhibition by the flavonoid natural product trans-chalcone. Chem. Commun. 2015, 51, 10483–10485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Becke, A.D. Density-functional thermochemistry. III. The role of exact exchange. J. Chem. Phys. 1993, 98, 5648–5652. [Google Scholar] [CrossRef] [Green Version]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A.; Nakatsuji, H.; et al. Gaussian 16 Revision 16.A.03; Wallingford CT. Inc.: Wallingford CT, UK, 2016. [Google Scholar]
- Thomsen, R.; Christensen, M.H. MolDock: A new technique for high accuracy molecular docking. J. Med. Chem. 2006, 49, 3315–3321. [Google Scholar] [CrossRef]
- Vale, V.V.; Cruz, J.N.; Viana, G.M.R.; Póvoa, M.M.; Brasil, D.d.S.B.; Dolabela, M.F. Naphthoquinones isolated from Eleutherine plicata herb: In vitro antimalarial activity and molecular modeling to investigate their binding modes. Med. Chem. Res. 2020, 29, 487–494. [Google Scholar] [CrossRef]
- Wang, J.; Wolf, R.M.; Caldwell, J.W.; Kollman, P.A.; Case, D.A. Development and testing of a general amber force field. J. Comput. Chem. 2004, 25, 1157–1174. [Google Scholar] [CrossRef]
- Cornell, W.D.; Cieplak, P.; Bayly, C.I.; Kollman, P.A. Application of RESP Charges to Calculate Conformational Energies, Hydrogen Bond Energies, and Free Energies of Solvation. J. Am. Chem. Soc. 1993, 115, 9620–9631. [Google Scholar] [CrossRef]
- Dolinsky, T.J.; Nielsen, J.E.; McCammon, J.A.; Baker, N.A. PDB2PQR: An automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 2004, 32, 665–667. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Robertson, A.D.; Jensen, J.H. Very fast empirical prediction and rationalization of protein pK a values. Proteins Struct. Funct. Genet. 2005, 61, 704–721. [Google Scholar] [CrossRef] [PubMed]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Case, D.A.; Cheatham, T.E.; Darden, T.; Gohlke, H.; Luo, R.; Merz, K.M.; Onufriev, A.; Simmerling, C.; Wang, B.; Woods, R.J. The Amber biomolecular simulation programs. J. Comput. Chem. 2005, 26, 1668–1688. [Google Scholar] [CrossRef] [Green Version]
- Silva, S.G.; da Costa, R.A.; de Oliveira, M.S.; da Cruz, J.N.; Figueiredo, P.L.B.; Brasil, D.d.S.B.; Nascimento, L.D.; Chaves Neto, A.M.d.J.; de Carvalho Junior, R.N.; Andrade, E.H.d.A. Chemical profile of Lippia thymoides, evaluation of the acetylcholinesterase inhibitory activity of its essential oil, and molecular docking and molecular dynamics simulations. PLoS ONE 2019, 14, e0213393. [Google Scholar] [CrossRef] [Green Version]
- de Oliveira, M.S.; da Cruz, J.N.; Gomes Silva, S.; da Costa, W.A.; de Sousa, S.H.B.; Bezerra, F.W.F.; Teixeira, E.; da Silva, N.J.N.; de Aguiar Andrade, E.H.; de Jesus Chaves Neto, A.M.; et al. Phytochemical profile, antioxidant activity, inhibition of acetylcholinesterase and interaction mechanism of the major components of the Piper divaricatum essential oil obtained by supercritical CO2. J. Supercrit. Fluids 2019, 145, 74–84. [Google Scholar] [CrossRef]
- Darden, T.; York, D.; Pedersen, L. Particle mesh Ewald: An N·log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef] [Green Version]
- Ryckaert, J.P.; Ciccotti, G.; Berendsen, H.J.C. Numerical integration of the cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. J. Comput. Phys. 1977, 23, 327–341. [Google Scholar] [CrossRef] [Green Version]
- Lzaguirre, J.A.; Catarello, D.P.; Wozniak, J.M.; Skeel, R.D. Langevin stabilization of molecular dynamics. J. Chem. Phys. 2001, 114, 2090–2098. [Google Scholar] [CrossRef]
- Cruz, J.N.; Costa, J.F.S.; Khayat, A.S.; Kuca, K.; Barros, C.A.L.; Neto, A.M.J.C. Molecular dynamics simulation and binding free energy studies of novel leads belonging to the benzofuran class inhibitors of Mycobacterium tuberculosis Polyketide Synthase 13. J. Biomol. Struct. Dyn. 2019, 37, 1616–1627. [Google Scholar] [CrossRef] [PubMed]
- Ramos, R.S.; Macêdo, W.J.C.; Costa, J.S.; da Silva, C.H.; Rosa, J.M.C.; da Cruz, J.N.; de Oliveira, M.S.; de Aguiar Andrade, E.H.; e Silva, R.B.L.; Souto, R.N.P.; et al. Potential inhibitors of the enzyme acetylcholinesterase and juvenile hormone with insecticidal activity: Study of the binding mode via docking and molecular dynamics simulations. J. Biomol. Struct. Dyn. 2019, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Neves Cruz, J.; Santana de Oliveira, M.; Gomes Silva, S.; Pedro da Silva Souza Filho, A.; Santiago Pereira, D.; Lima e Lima, A.H.; de Aguiar Andrade, E.H. Insight into the Interaction Mechanism of Nicotine, NNK, and NNN with Cytochrome P450 2A13 Based on Molecular Dynamics Simulation. J. Chem. Inf. Model. 2020, 60, 766–776. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds (essential oil extracted from Siparuna guianensis Aublet) are available from the authors. |




| Rt | RIC | RIL | Compound | PubChem CID/SID or Chemical Structure | Concentration (%) |
|---|---|---|---|---|---|
| 5.817 | 933 | 932 a | α-pinene | 6654 | 0.33 |
| 7.034 | 978 | 974 a | β-pinene | 14896 | 0.04 |
| 7.375 | 989 | 988 a | myrcene | 31253 | 0.22 |
| 7.465 | 1008 | 1002 a | α-phellandrene | 7460 | 0.15 |
| 8.808 | 1030 | 1025 a | sylvestrene | 12304570 | 0.51 |
| 8.998 | 1044 | 1044 a | (E)-β-ocimene | 5281553 | 0.07 |
| 19.958 | 1292 | 1293 a | undecan-2-one | 8163 | 0.38 |
| 21.842 | 1331 | 1335 a | δ-elemene | 12309449 | 5.38 |
| 22.342 | 1345 | 1345 a | α-cubebene | 86609 | 0.48 |
| 23.317 | 1367 | 1373 a | α-ylangene | 442409 | 0.12 |
| 23.608 | 1373 | 1374 a | α-copaene | 442355 | 1.1 |
| 23.879 | 1381 | 1387 a | β-bourbonene | 62566 | 0.52 |
| 23.942 | 1383 | 1389 a | β-elemene | 6918391 | 0.55 |
| 24.258 | 1386 | 1387 a | β-Cubebene | 93081 | 3.34 |
| 24.654 | 1392 | 1402 a | α-funebrene | 6552024 | 0.03 |
| 24.967 | 1404 | 1409 a | α-gurjunene | 15560276 | 0.06 |
| 25.158 | 1408 | 1417 a | (E)-caryophyllene | 5281515 | 0.03 |
| 25.525 | 1417 | 1419 a | β-ylangene | 519779 | 4.14 |
| 26.1 | 1430 | 1434 a | γ-elemene | 6432312 | 7.04 |
| 26.242 | 1434 | 1437 a | α-guaiene | 5317844 | 0.23 |
| 26.375 | 1437 | 1439 a | aromadendrene | 91354 | 0.19 |
| 26.483 | 1439 | 1442 a | guaia-6,9-diene | 6427475 | 0.12 |
| 26.883 | 1449 | 1448 a | cis-muurola-3,5-diene | 51351708 | 1.4 |
| 27.075 | 1453 | 1452 | α-humulene | 5281520 | 0.86 |
| 27.408 | 1457 | 1458 a | alloaromadendrene | 10899740 | 0.29 |
| 27.608 | 1459 | 1461 a | cis-cadina-1(6),4-diene | 6431126 | 0.35 |
| 27.788 | 1466 | 1464 a | 9-epi-(E)-caryophyllene | 6429274 | 0.09 |
| 27.892 | 1471 | 1475 a | γ-gurjunene | 90805 | 0.49 |
| 27.925 | 1475 | 1475 a | γ-muurolene | 12313020 | 0.7 |
| 28.325 | 1482 | 1480 a | germacrene D | 5373727 | 7.61 |
| 28.55 | 1488 | 1489 a | β-selinene | 442393 | 1.61 |
| 28.642 | 1490 | 1493 a | trans-muurola-4(14),5-diene | 91747125 | 0.63 |
| 28.892 | 1496 | 1499 a | curzerene | 572766 | 7.1 |
| 28.992 | 1498 | 1500 a | α-muurolene | 12306047 | 1.2 |
| 29.125 | 1501 | 1495 a | γ-amorphene | 12313019 | 0.48 |
| 29.467 | 1506 | 1508 a | germacrene A | 9548705 | 0.02 |
| 29.525 | 1511 | 1513 a | γ-cadinene | 92313 | 0.39 |
| 29.758 | 1516 | 1522 a | δ-cadinene | 12306054 | 1.86 |
| 29.925 | 1521 | 1528 a | zonarene | 6428488 | 0.48 |
| 30.317 | 1530 | 1533 a | trans-cadina-1,4-diene | 91746579 | 0.45 |
| 30.492 | 1534 | 1531 b | selina-4(14),7(11)-diene | 10655819 | 1.04 |
| 30.667 | 1539 | 1545 a | selina-3,7(11)-diene | 522296 | 0.25 |
| 31.375 | 1556 | 1559 a | germacrene B | 5281519 | 1.88 |
| 32.308 | 1582 | 1589 a | allo-hedycaryol |  [46] [46] | 0.51 |
| 32.445 | 1592 | 1589 a | cis-β-elemenone | 519762 | 1.43 |
| 33.258 | 1602 | 1602 a | trans-β-elemenone |  [47] [47] | 11.78 |
| 34.492 | 1633 | 1627 a | cubenol<1-epi-> | 91753500 | 0.97 |
| 34.583 | 1643 | 1645 a | cubenol | 1770062 | 1.15 |
| 35.867 | 1661 | 1657 a | atractylone | 3080635 | 18.65 |
| 36.85 | 1694 | 1693 a | germacrone | 5317571 | 5.26 |
| 38.375 | 1739 | 1740 a | mint sulfide | 14564587 | 0.05 |
| Monoterpene Hydrocarbons | 1.35 | ||||
| Sesquiterpene Hydrocarbons | 47.84 | ||||
| Oxygenated Sesquiterpenes | 46.85 | ||||
| Others | 0.43 | ||||
| Total | 96.47 | ||||
| Sample/ Dilution (µL/mL) | A | B | C | d | |
|---|---|---|---|---|---|
| 1 | 500 | - | - | MIC | - |
| 2 | 250 | - | MIC | + | - |
| 3 | 125 | MIC | + | + | MIC |
| 4 | 62.5 | + | + | + | + |
| 5 | 30.625 | + | + | + | + |
| 6 | 15.3 | + | + | + | + |
| 7 | 7.6 | + | + | + | + |
| 8 | 3.8 | + | + | + | + |
| 9 | 1.9 | + | + | + | + |
| 10 | 0.95 | + | + | + | + |
| Mean halo, 10 µL, N = 3 | 11 ± 0.12 | 12 ± 0.57 | 11 ± 0,31 | 12.5 ± 0,98 | |
| Control | 22.5 ± 0.32 | 28.10 ± 0.13 | 15.25 ± 0.58 | 19.42 ± 1.22 | |
| Targets | MolDock Score |
|---|---|
| C. albicans | −71.43 |
| E. coli | −87.24 |
| E. faecalis | −80.46 |
| S. mutans | −65.18 |
| Targets | ΔEvdW | ΔEele | ΔGGB | ΔGNP | ΔGMM-GBSA |
|---|---|---|---|---|---|
| C. albicans | −22.28 | −5.51 | 13.74 | −13.11 | −25.16 |
| E. coli | −25.54 | −6.88 | 15.96 | −9.87 | −26.33 |
| E. faecalis | −19.56 | −5.02 | 8.96 | −8.22 | −23.84 |
| S. mutans | −24.35 | −3.74 | 9.75 | −9.13 | −27.47 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santana de Oliveira, M.; da Cruz, J.N.; Almeida da Costa, W.; Silva, S.G.; Brito, M.d.P.; de Menezes, S.A.F.; de Jesus Chaves Neto, A.M.; de Aguiar Andrade, E.H.; de Carvalho Junior, R.N. Chemical Composition, Antimicrobial Properties of Siparuna guianensis Essential Oil and a Molecular Docking and Dynamics Molecular Study of its Major Chemical Constituent. Molecules 2020, 25, 3852. https://doi.org/10.3390/molecules25173852
Santana de Oliveira M, da Cruz JN, Almeida da Costa W, Silva SG, Brito MdP, de Menezes SAF, de Jesus Chaves Neto AM, de Aguiar Andrade EH, de Carvalho Junior RN. Chemical Composition, Antimicrobial Properties of Siparuna guianensis Essential Oil and a Molecular Docking and Dynamics Molecular Study of its Major Chemical Constituent. Molecules. 2020; 25(17):3852. https://doi.org/10.3390/molecules25173852
Chicago/Turabian StyleSantana de Oliveira, Mozaniel, Jorddy Neves da Cruz, Wanessa Almeida da Costa, Sebastião Gomes Silva, Mileide da Paz Brito, Sílvio Augusto Fernandes de Menezes, Antônio Maia de Jesus Chaves Neto, Eloisa Helena de Aguiar Andrade, and Raul Nunes de Carvalho Junior. 2020. "Chemical Composition, Antimicrobial Properties of Siparuna guianensis Essential Oil and a Molecular Docking and Dynamics Molecular Study of its Major Chemical Constituent" Molecules 25, no. 17: 3852. https://doi.org/10.3390/molecules25173852
APA StyleSantana de Oliveira, M., da Cruz, J. N., Almeida da Costa, W., Silva, S. G., Brito, M. d. P., de Menezes, S. A. F., de Jesus Chaves Neto, A. M., de Aguiar Andrade, E. H., & de Carvalho Junior, R. N. (2020). Chemical Composition, Antimicrobial Properties of Siparuna guianensis Essential Oil and a Molecular Docking and Dynamics Molecular Study of its Major Chemical Constituent. Molecules, 25(17), 3852. https://doi.org/10.3390/molecules25173852