Docking Prediction, Antifungal Activity, Anti-Biofilm Effects on Candida spp., and Toxicity against Human Cells of Cinnamaldehyde
Abstract
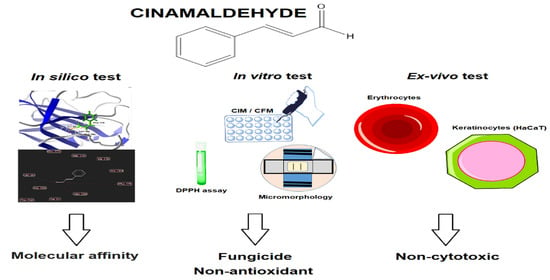
:1. Introduction
2. Results
2.1. Molecular Dockings
2.2. Determination of the Minimum Inhibitory Concentration and Minimum Fungicidal Concentration
2.3. Sorbitol and Ergosterol Assays
2.4. Effects of Cinnamaldehyde on Fungal Micromorphology
2.5. Antibiofilm Activity of Cinnamaldehyde
2.6. Cytotoxicity of Cinnamaldehyde in Keratinocytes
2.7. Cytotoxicity in Human Erythrocytes
2.8. Antioxidant Activity of Cinnamaldehyde by the DPPH Method
3. Discussion
4. Materials and Methods
4.1. Molecular Docking
4.2. Chemicals and Microorganisms
4.3. Determination of the Minimum Inhibitory Concentration (MIC) and Minimum Fungicidal Concentration (MFC)
4.4. Effects of Cinnamaldehyde on the Fungal Cell Wall and Membrane Permeability
4.4.1. Sorbitol Test (Effect on Cell Wall)
4.4.2. Ergosterol Test (Effect on Cell Membrane)
4.5. Effects of Cinnamaldehyde on Fungal Micromorphology
4.6. Effects of Cinnamaldehyde on Biofilm Reduction
4.7. MTT Cell Viability Assay
4.8. Cytotoxic Effects of Cinnamaldehyde on Human Erythrocytes
4.9. Antioxidant Activity of Cinnamaldehyde by the DPPH Method
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Singh, A.; Verma, R.; Murari, A.; Agrawal, A. Oral Candidiasis: An Overview. J. Oral Maxillofac. Pathol. JOMFP 2014, 18, S81. [Google Scholar]
- Billings, M.; Dye, B.A.; Iafolla, T.; Grisius, M.; Alevizos, I. Elucidating the Role of Hyposalivation and Autoimmunity in Oral Candidiasis. Oral Dis. 2017, 23, 387–394. [Google Scholar] [CrossRef] [PubMed]
- Muadcheingka, T.; Tantivitayakul, P. Distribution of Candida albicans and Non-albicans Candida Species in Oral Candidiasis Patients: Correlation between Cell Surface Hydrophobicity and Biofilm Forming Activities. Arch. Oral Biol. 2015, 60, 894–901. [Google Scholar] [CrossRef]
- Patil, S.; Rao, R.S.; Majumdar, B.; Anil, S. Clinical Appearance of Oral Candida Infection and Therapeutic Strategies. Front. Microbiol. 2015, 6, 1391. [Google Scholar] [CrossRef] [Green Version]
- Manik, A.; Bahl, R. A Review on Oral Candidal Infection. J. Adv. Med. Dent. Sci. Res. 2017, 5, 54. [Google Scholar]
- Junqueira, J.C.; Vilela, S.F.G.; Rossoni, R.D.; Barbosa, J.O.; Costa, A.C.B.P.; Rasteiro, V.; Suleiman, J.M.A.H.; Jorge, A.O.C. Oral Colonization by Yeasts in HIV-Positive Patients in Brazil. Rev. Inst. Med. Trop. Sao Paulo 2012, 54, 17–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fox, E.P.; Nobile, C.J. Candida albicans: Symptoms, Causes and Treatment Options; Dietrich, L.A., Friedmann, T.S., Eds.; Nova Science Publishers: New York, NY, USA, 2013. [Google Scholar]
- Doddanna, S.J.; Patel, S.; Sundarrao, M.A.; Veerabhadrappa, R.S. Antimicrobial Activity of Plant Extracts on Candida albicans: An in vitro Study. Indian J. Dent. Res. 2013, 24, 401. [Google Scholar] [CrossRef] [PubMed]
- Barbosa, M.B.; Faria, M.G.I. Produtos Naturais Como Nova Alternativa Terapêutica Para o Tratamento de Candidíase Bucal. Rev. Uningá Rev. 2014, 20, 103–107. [Google Scholar]
- Arendrup, M.C.; Patterson, T.F. Multidrug-Resistant Candida: Epidemiology, Molecular Mechanisms, and Treatment. J. Infect. Dis. 2017, 216, S445–S451. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Del Valle, G.M.M. Candida Glabrata: Un Patógeno Emergente. Biociencias 2015, 10, 89–102. [Google Scholar] [CrossRef]
- El-Houssaini, H.H.; Elnabawy, O.M.; Nasser, H.A.; Elkhatib, W.F. Influence of Subinhibitory Antifungal Concentrations on Extracellular Hydrolases and Biofilm Production by Candida albicans Recovered from Egyptian Patients. BMC Infect. Dis. 2019, 19, 54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perlin, D.S.; Rautemaa-Richardson, R.; Alastruey-Izquierdo, A. The Global Problem of Antifungal Resistance: Prevalence, Mechanisms, and Management. Lancet Infect. Dis. 2017, 17, e383–e392. [Google Scholar] [CrossRef]
- Robbins, N.; Caplan, T.; Cowen, L.E. Molecular Evolution of Antifungal Drug Resistance. Annu. Rev. Microbiol. 2017, 71, 753–775. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simões, C.M.O.; Schenkel, E.P.; de Mello, J.C.P.; Mentz, L.A.; Petrovick, P.R. Farmacognosia: Do Produto Natural Ao Medicamento; Artmed Editora: Porto Alegre, Brazil, 2016. [Google Scholar]
- Khasnavis, S.; Pahan, K. Sodium Benzoate, a Metabolite of Cinnamon and a Food Additive, Upregulates Neuroprotective Parkinson Disease Protein DJ-1 in Astrocytes and Neurons. J. Neuroimmune Pharmacol. 2012, 7, 424–435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malik, J.; Munjal, K.; Deshmukh, R. Attenuating Effect of Standardized Lyophilized Cinnamomum Zeylanicum Bark Extract against Streptozotocin-Induced Experimental Dementia of Alzheimer’s Type. J. Basic Clin. Physiol. Pharmacol. 2015, 26, 275–285. [Google Scholar] [CrossRef] [PubMed]
- Chung, J.-W.; Kim, J.-J.; Kim, S.-J. Antioxidative Effects of Cinnamomi Cortex: A Potential Role of INOS and COX-II. Pharmacogn. Mag. 2011, 7, 314. [Google Scholar]
- Mollazadeh, H.; Hosseinzadeh, H. Cinnamon Effects on Metabolic Syndrome: A Review Based on Its Mechanisms. Iran. J. Basic Med. Sci. 2016, 19, 1258. [Google Scholar]
- Hamidpour, R.; Hamidpour, M.; Hamidpour, S.; Shahlari, M. Cinnamon from the Selection of Traditional Applications to Its Novel Effects on the Inhibition of Angiogenesis in Cancer Cells and Prevention of Alzheimer’s Disease, and a Series of Functions Such as Antioxidant, Anticholesterol, Antidiabetes, Antibacteri. J. Tradit. Complement. Med. 2015, 5, 66–70. [Google Scholar] [CrossRef] [Green Version]
- Connell, B.J.; Chang, S.-Y.; Prakash, E.; Yousfi, R.; Mohan, V.; Posch, W.; Wilflingseder, D.; Moog, C.; Kodama, E.N.; Clayette, P. A Cinnamon-Derived Procyanidin Compound Displays Anti-HIV-1 Activity by Blocking Heparan Sulfate-and Co-Receptor-Binding Sites on Gp120 and Reverses T Cell Exhaustion via Impeding Tim-3 and PD-1 Upregulation. PLoS ONE 2016, 11, e0165386. [Google Scholar] [CrossRef]
- Kim, J.; Bao, T.H.Q.; Shin, Y.-K.; Kim, K.-Y. Antifungal Activity of Magnoflorine against Candida Strains. World J. Microbiol. Biotechnol. 2018, 34, 167. [Google Scholar] [CrossRef]
- Shreaz, S.; Sheikh, R.A.; Rimple, B.; Hashmi, A.A.; Nikhat, M.; Khan, L.A. Anticandidal Activity of Cinnamaldehyde, Its Ligand and Ni (II) Complex: Effect of Increase in Ring and Side Chain. Microb. Pathog. 2010, 49, 75–82. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.S.A.; Ahmad, I. Antibiofilm Activity of Certain Phytocompounds and Their Synergy with Fluconazole against Candida albicans Biofilms. J. Antimicrob. Chemother. 2011, 67, 618–621. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, S.N.; Khan, S.; Iqbal, J.; Khan, R.; Khan, A.U. Enhanced Killing and Antibiofilm Activity of Encapsulated Cinnamaldehyde against Candida albicans. Front. Microbiol. 2017, 8, 1641. [Google Scholar] [CrossRef] [PubMed]
- Shreaz, S.; Bhatia, R.; Khan, N.; Muralidhar, S.; Basir, S.F.; Manzoor, N.; Khan, L.A. Spice Oil Cinnamaldehyde Exhibits Potent Anticandidal Activity against Fluconazole Resistant Clinical Isolates. Fitoterapia 2011, 82, 1012–1020. [Google Scholar] [CrossRef] [PubMed]
- Silva, D.R.; Sardi, J.D.C.O.; Freires, I.A.; Silva, A.C.B.; Rosalen, P.L. In Silico Approaches for Screening Molecular Targets in Candida albicans: A Proteomic Insight into Drug Discovery and Development. Eur. J. Pharmacol. 2019, 842, 64–69. [Google Scholar] [CrossRef]
- Jovanović, M.; Obradović, R.; Pejčić, A.; Stanišić, D.; Stošić, N.; Popović, Ž. The Role of Candida albicans on the Development of Stomatitis in Patients Wearing Dentures. Sanamed 2018, 13, 175–182. [Google Scholar] [CrossRef] [Green Version]
- Taudorf, E.H.; Jemec, G.B.E.; Hay, R.J.; Saunte, D.M.L. Cutaneous Candidiasis–an Evidence-based Review of Topical and Systemic Treatments to Inform Clinical Practice. J. Eur. Acad. Dermatology Venereol. 2019, 33, 1863–1873. [Google Scholar] [CrossRef]
- Gow, N.A.R.; Latge, J.-P.; Munro, C.A. The Fungal Cell Wall: Structure, Biosynthesis, and Function. Fungal Kingd. 2017, 267–292. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.-F.; Xia, J.-J.; Nie, K.-L.; Wang, F.; Deng, L. Outline of the Biosynthesis and Regulation of Ergosterol in Yeast. World J. Microbiol. Biotechnol. 2019, 35, 98. [Google Scholar] [CrossRef]
- Choi, M.; Karunaratne, K.; Kohen, A. Flavin-Dependent Thymidylate Synthase as a New Antibiotic Target. Molecules 2016, 21, 654. [Google Scholar] [CrossRef] [Green Version]
- Pootong, A.; Norrapong, B.; Cowawintaweewat, S. Antifungal Activity of Cinnamaldehyde against Candida albicans. Southeast Asian J. Trop. Med. Public Health 2017, 48, 150–158. [Google Scholar] [PubMed]
- Wayne, P. Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts; Approved Standard; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2008. [Google Scholar]
- Freire, J.C.P.; Júnior, J.K.D.O.; Silva, D.D.F.; Sousa, J.P.D.; Guerra, F.Q.S.; de Oliveira Lima, E. Antifungal Activity of Essential Oils against Candida albicans Strains Isolated from Users of Dental Prostheses. Evid. Based Complement. Altern. Med. 2017, 2017, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scheibler, E.; Garcia, M.C.R.; Medina da Silva, R.; Figueiredo, M.A.; Salum, F.G.; Cherubini, K. Use of Nystatin and Chlorhexidine in Oral Medicine: Properties, Indications and Pitfalls with Focus on Geriatric Patients. Gerodontology 2017, 34, 291–298. [Google Scholar] [CrossRef] [PubMed]
- Shreaz, S.; Bhatia, R.; Khan, N.; Muralidhar, S.; Manzoor, N.; Khan, L.A. Influences of Cinnamic Aldehydes on H+ Extrusion Activity and Ultrastructure of Candida. J. Med. Microbiol. 2013, 62, 232–240. [Google Scholar] [CrossRef] [Green Version]
- Shreaz, S.; Wani, W.A.; Behbehani, J.M.; Raja, V.; Irshad, M.; Karched, M.; Ali, I.; Siddiqi, W.A.; Hun, L.T. Cinnamaldehyde and Its Derivatives, a Novel Class of Antifungal Agents. Fitoterapia 2016, 112, 116–131. [Google Scholar] [CrossRef]
- Hu, L.; Wang, D.; Liu, L.; Chen, J.; Xue, Y.; Shi, Z. Ca2+ Efflux Is Involved in Cinnamaldehyde-Induced Growth Inhibition of Phytophthora Capsici. PLoS ONE 2013, 8, e76264. [Google Scholar] [CrossRef] [Green Version]
- Khan, M.S.A.; Ahmad, I.; Cameotra, S.S. Phenyl Aldehyde and Propanoids Exert Multiple Sites of Action towards Cell Membrane and Cell Wall Targeting Ergosterol in Candida albicans. Amb Express 2013, 3, 54. [Google Scholar] [CrossRef] [Green Version]
- Rajput, S.B.; Karuppayil, S.M. Small Molecules Inhibit Growth, Viability and Ergosterol Biosynthesis in Candida albicans. Springerplus 2013, 2, 26. [Google Scholar] [CrossRef] [Green Version]
- Yossa, N.; Patel, J.; Millner, P.; Ravishankar, S.; Lo, Y.M. Antimicrobial Activity of Plant Essential Oils against Escherichia Coli O157: H7 and Salmonella on Lettuce. Foodborne Pathog. Dis. 2013, 10, 87–96. [Google Scholar] [CrossRef]
- Ferreira, G.; Rosalen, P.; Peixoto, L.; Pérez, A.; Carlo, F.; Castellano, L.; Lima, J.; Freires, I.; Lima, E.; Castro, R. Antibiofilm Activity and Mechanism of Action of the Disinfectant Chloramine T on Candida Spp., and Its Toxicity against Human Cells. Molecules 2017, 22, 1527. [Google Scholar] [CrossRef] [Green Version]
- Taguchi, Y.; Hasumi, Y.; Abe, S.; Nishiyama, Y. The Effect of Cinnamaldehyde on the Growth and the Morphology of Candida albicans. Med. Mol. Morphol. 2013, 46, 8–13. [Google Scholar] [CrossRef] [PubMed]
- Pakkulnan, R.; Anutrakunchai, C.; Kanthawong, S.; Taweechaisupapong, S.; Chareonsudjai, P.; Chareonsudjai, S. Extracellular DNA Facilitates Bacterial Adhesion during Burkholderia Pseudomallei Biofilm Formation. PLoS ONE 2019, 14, e0213288. [Google Scholar] [CrossRef] [PubMed]
- Skowron, K.; Wiktorczyk, N.; Grudlewska, K.; Kwiecińska-Piróg, J.; Wałecka-Zacharska, E.; Paluszak, Z.; Gospodarek-Komkowska, E. Drug-Susceptibility, Biofilm-Forming Ability and Biofilm Survival on Stainless Steel of Listeria Spp. strains Isolated from Cheese. Int. J. Food Microbiol. 2019, 296, 75–82. [Google Scholar] [CrossRef] [PubMed]
- Katchburian, E.; Arana Chavez, V.E. Histologia e Embriologia Oral: Texto, Atlas, Correlações Clínicas; Guanabara Koogan: Rio de Janeiro, Brazil, 2012. [Google Scholar]
- García-Salinas, S.; Elizondo-Castillo, H.; Arruebo, M.; Mendoza, G.; Irusta, S. Evaluation of the Antimicrobial Activity and Cytotoxicity of Different Components of Natural Origin Present in Essential Oils. Molecules 2018, 23, 1399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- ISO 10993-5. Biological Evaluation of Medical Devices—Part 5: Tests for Cytotoxicity: In Vitro Methods; ISO: Geneva, Switzerland, 1992. [Google Scholar]
- Yu, C.; Liu, S.-L.; Qi, M.-H.; Zou, X. Cinnamaldehyde/Chemotherapeutic Agents Interaction and Drug-Metabolizing Genes in Colorectal Cancer. Mol. Med. Rep. 2014, 9, 669–676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Divkovic, M.; Pease, C.K.; Gerberick, G.F.; Basketter, D.A. Hapten–Protein Binding: From Theory to Practical Application in the in vitro Prediction of Skin Sensitization. Contact Dermat. 2005, 53, 189–200. [Google Scholar] [CrossRef]
- Suwalsky, M.; Vargas, P.; Avello, M.; Villena, F.; Sotomayor, C.P. Human Erythrocytes Are Affected in vitro by Flavonoids of Aristotelia Chilensis (Maqui) Leaves. Int. J. Pharm. 2008, 363, 85–90. [Google Scholar] [CrossRef]
- Dobrovolskaia, M.A.; McNeil, S.E. Understanding the Correlation between in vitro and in Vivo Immunotoxicity Tests for Nanomedicines. J. Control. Release 2013, 172, 456–466. [Google Scholar] [CrossRef] [Green Version]
- Farag, M.R.; Alagawany, M. Erythrocytes as a Biological Model for Screening of Xenobiotics Toxicity. Chem. Biol. Interact. 2018, 279, 73–83. [Google Scholar] [CrossRef]
- Scherer, R.; Wagner, R.; Duarte, M.C.T.; Godoy, H.T. Composição e Atividades Antioxidante e Antimicrobiana Dos Óleos Essenciais de Cravo-Da-Índia, Citronela e Palmarosa. Rev. Bras. Plantas Med. 2009, 11, 442–449. [Google Scholar] [CrossRef]
- Release, H. 7.5 for Windows, Molecular Modeling System; Hypercube Inc.: Waterloo, ON, Canada, 2002. [Google Scholar]
- De Campos, L.V.B.; Correia, J.C.G.; Carauta, A.N.M. Estudo Da Interação Do Trietoxisilano Com o Ácido Linoléico Como Hidrofugante Em Rochas Ornamentais via Modelagem Molecular; CETEM/MCTIC: São Paulo, Brazil, 2017.
- Moreira, M.P. Novos Polímeros a Base de Ácido Glicerofosfórico/Beta-Ciclodextrina Reticulado Com Ligações Uretânicas: Preparação e Incorporação de Ciprofloxacina; Universidade Federal de Sergipe: Sergipe, Brazil, 2017. [Google Scholar]
- Altê, M.A. Estudo Estrutural e Planejamento de Novos Inibidores Para a Enzima Prefenato Desidratase de Mycobacterium Tuberculosis; Universidade Federal de Ciências da Saúde de Porto: Alegre, Brazil, 2017. [Google Scholar]
- Barros, R.P.C. Triagem Virtual de Metabólitos Secundários Com Potencial Atividade Antimicrobiana Do Gênero Solanum e Estudo Fitoquimico de Solanum Capsicoides All; Universidade Federal da Paraíba: João Pessoa, Brazil, 2017. [Google Scholar]
- Santana, C.B. Composição Química, Atividade Antimicrobiana, Inseticida e Antioxidante Do Óleo Essencial e Extratos de Myrcia Oblongata DC; Universidade Estadual do Oeste do Paraná: Cascavel, Brazil, 2017. [Google Scholar]
- Hargrove, T.Y.; Friggeri, L.; Wawrzak, Z.; Qi, A.; Hoekstra, W.J.; Schotzinger, R.J.; York, J.D.; Guengerich, F.P.; Lepesheva, G.I. Structural Analyses of Candida albicans Sterol 14α-Demethylase Complexed with Azole Drugs Address the Molecular Basis of Azole-Mediated Inhibition of Fungal Sterol Biosynthesis. J. Biol. Chem. 2017, 292, 6728–6743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Roberti, R.; Blobel, G. Structure of an Integral Membrane Sterol Reductase from Methylomicrobium alcaliphilum. Nature 2015, 517, 104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cutfield, J.F.; Sullivan, P.A.; Cutfield, S.M. Minor Structural Consequences of Alternative CUG Codon Usage (Ser for Leu) in Candida albicans Exoglucanase. Protein Eng. 2000, 13, 735–738. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sinha, K.; Rule, G.S. The Structure of Thymidylate Kinase from Candida albicans Reveals a Unique Structural Element. Biochemistry 2017, 56, 4360–4370. [Google Scholar] [CrossRef] [PubMed]
- Hemsworth, G.R.; Henrissat, B.; Davies, G.J.; Walton, P.H. Discovery and Characterization of a New Family of Lytic Polysaccharide Monooxygenases. Nat. Chem. Biol. 2014, 10, 122. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.E.; Chehri, K.; Karimi, N.; Karimi, I. Computational Approaches to the in vitro Antibacterial Activity of Allium airtifolium Boiss against Gentamicin-Resistant Escherichia Coli: Focus on Ribosome Recycling Factor. Silico Pharmacol. 2017, 5, 7. [Google Scholar] [CrossRef] [Green Version]
- Varma, P.B.S.; Adimulam, Y.B.; Subrahmanyam, K. In Silico Virtual Screening of PubChem Compounds against Phosphotransacetylase, a Putative Drug Target for Staphylococcus aureus. Int. J. Comput. Biol. Drug Des. 2017, 10, 39–48. [Google Scholar] [CrossRef]
- Wu, Y.-Y.; Zhang, T.-Y.; Zhang, M.-Y.; Cheng, J.; Zhang, Y.-X. An Endophytic Fungi of Ginkgo Biloba L. Produces Antimicrobial Metabolites as Potential Inhibitors of FtsZ of Staphylococcus aureus. Fitoterapia 2018, 128, 265–271. [Google Scholar]
- Loo, J.S.E.; Emtage, A.L.; Ng, K.W.; Yong, A.S.J.; Doughty, S.W. Assessing GPCR Homology Models Constructed from Templates of Various Transmembrane Sequence Identities: Binding Mode Prediction and Docking Enrichment. J. Mol. Graph. Model. 2018, 80, 38–47. [Google Scholar] [CrossRef]
- Ounthaisong, U.; Tangyuenyongwatana, P. Cross-Docking Study of Flavonoids against Tyrosinase Enzymes Using PyRx 0.8 Virtual Screening Tool. TJPS 2017, 41, 189–192. [Google Scholar]
- Wang, T.; Yang, Z.; Zhang, Y.; Yan, W.; Wang, F.; He, L.; Zhou, Y.; Chen, L. Discovery of Novel CDK8 Inhibitors Using Multiple Crystal Structures in Docking-Based Virtual Screening. Eur. J. Med. Chem. 2017, 129, 275–286. [Google Scholar] [CrossRef] [PubMed]
- Yanuar, A.; Pratiwi, I.; Syahdi, R.R. In Silico Activity Analysis of Saponins and 2, 5-Piperazinedione from Marine Organism against Murine Double Minute-2 Inhibitor and Procaspase-3 Activator. J. Young Pharm. 2018, 10, S16. [Google Scholar] [CrossRef] [Green Version]
- Rasooli, I.; Abyaneh, M.R. Inhibitory Effects of Thyme Oils on Growth and Aflatoxin Production by Aspergillus Parasiticus. Food Control 2004, 15, 479–483. [Google Scholar] [CrossRef]
- Siddiqui, Z.N.; Farooq, F.; Musthafa, T.N.M.; Ahmad, A.; Khan, A.U. Synthesis, Characterization and Antimicrobial Evaluation of Novel Halopyrazole Derivatives. J. Saudi Chem. Soc. 2013, 17, 237–243. [Google Scholar] [CrossRef] [Green Version]
- Frost, D.J.; Brandt, K.I.M.D.; Cugier, D.; Goldman, R. A Whole-Cell Candida albicans Assay for the Detection of Inhibitors towards Fungal Cell Wall Synthesis and Assembly. J. Antibiot. 1995, 48, 306–310. [Google Scholar] [CrossRef] [Green Version]
- Leite, M.C.A.; Bezerra, A.P.D.B.; Sousa, J.P.D.; Guerra, F.Q.S.; Lima, E.D.O. Evaluation of Antifungal Activity and Mechanism of Action of Citral against Candida albicans. Evid. Based Complement. Altern. Med. 2014, 2014, 1–9. [Google Scholar] [CrossRef] [Green Version]
- De Almeida Freires, I.; Murata, R.M.; Furletti, V.F.; Sartoratto, A.; de Alencar, S.M.; Figueira, G.M.; de Oliveira Rodrigues, J.A.; Duarte, M.C.T.; Rosalen, P.L. Coriandrum sativum L.(Coriander) Essential Oil: Antifungal Activity and Mode of Action on Candida Spp., and Molecular Targets Affected in Human Whole-Genome Expression. PLoS ONE 2014, 9, e99086. [Google Scholar]
- Lima, I.O.; Pereira, F.D.; Oliveira, W.A.; Lima, E.D.; Menezes, E.A.; Cunha, F.A.; Diniz, M.D. Antifungal Activity and Mode of Action of Carvacrol against Candida albicans Strains. J. Essent. Oil Res. 2013, 25, 138–142. [Google Scholar] [CrossRef]
- Hao, B.; Cheng, S.; Clancy, C.J.; Nguyen, M.H. Caspofungin Kills Candida albicans by Causing Both Cellular Apoptosis and Necrosis. Antimicrob. Agents Chemother. 2013, 57, 326–332. [Google Scholar] [CrossRef] [Green Version]
- Letscher-Bru, V.; Herbrecht, R. Caspofungin: The First Representative of a New Antifungal Class. J. Antimicrob. Chemother. 2003, 51, 513–521. [Google Scholar] [CrossRef] [Green Version]
- Pierce, C.G.; Srinivasan, A.; Uppuluri, P.; Ramasubramanian, A.K.; Lopez-Ribot, J.L. Antifungal Therapy with an Emphasis on Biofilms. Curr. Opin. Pharmacol. 2013, 13, 726–730. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ellepola, A.N.B.; Samaranayake, L.P. Impact of Brief and Sequential Exposure to Nystatin on the Germ Tube Formation and Cell Surface Hydrophobicity of Oral Candida albicans Isolates from Human Immunodeficiency Virus-Infected Patients. Med. Princ. Pract. 2014, 23, 307–312. [Google Scholar] [CrossRef] [PubMed]
- Alves, L.A.; Freires, I.D.; Pereira, T.M.; Souza, A.D.; Lima, E.D.; Castro, R.D. Effect of Schinus Terebinthifolius on Candida albicans Growth Kinetics, Cell Wall Formation and Micromorphology. Acta Odontol. Scand. 2013, 71, 965–971. [Google Scholar] [CrossRef] [PubMed]
- Ellepola, K.; Liu, Y.; Cao, T.; Koo, H.; Seneviratne, C.J. Bacterial GtfB Augments Candida albicans Accumulation in Cross-Kingdom Biofilms. J. Dent. Res. 2017, 96, 1129–1135. [Google Scholar] [CrossRef] [PubMed]
- Denizot, F.; Lang, R. Rapid Colorimetric Assay for Cell Growth and Survival: Modifications to the Tetrazolium Dye Procedure Giving Improved Sensitivity and Reliability. J. Immunol. Methods 1986, 89, 271–277. [Google Scholar] [CrossRef]
- Jain, K.; Verma, A.K.; Mishra, P.R.; Jain, N.K. Surface-Engineered Dendrimeric Nanoconjugates for Macrophage-Targeted Delivery of Amphotericin B: Formulation Development and in vitro and in Vivo Evaluation. Antimicrob. Agents Chemother. 2015, 59, 2479–2487. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brand-Williams, W.; Cuvelier, M.-E.; Berset, C. Use of a Free Radical Method to Evaluate Antioxidant Activity. LWT Food Sci. Technol. 1995, 28, 25–30. [Google Scholar] [CrossRef]
- Kim, D.-O.; Lee, K.W.; Lee, H.J.; Lee, C.Y. Vitamin C Equivalent Antioxidant Capacity (VCEAC) of Phenolic Phytochemicals. J. Agric. Food Chem. 2002, 50, 3713–3717. [Google Scholar] [CrossRef] [PubMed]





| ID | 1EQP | 4MAI | 4QUV | 5TZ1 | 5UIV |
|---|---|---|---|---|---|
| Cinnamaldehyde | −61.1458 | −70.4951 | −59.8535 | −51.4852 | −66.4852 |
| Miconazole | −135.5710 | 86.0729 | −96.3498 | −79.7099 | −110.4430 |
| Nystatin | −100.6060 | 663.2230 | 21.5918 | 168.3950 | 140.0100 |
| Ligand | - | - | −203.0010 | −81.7980 | −148.3980 |
| RMSD | - | - | 0.2048 | 0.4409 | 0.2112 |
| Hydrogen Bonds (amount) | Tyr 29 (1) Tyr 255 (1) | Gln 66 (1) | Arg 323 (1) Tyr 407 (1) | Arg 92 (1) | |
| Electrostatic Interactions | - | - | - | ||
| Steric Interactions | Asp 145 (1) Trp 363 (1) | Phe 175 (1) Asn 194 (2) | - | Net 508 (1) |
| Cinnamaldehyde | Nystatin | |||||
|---|---|---|---|---|---|---|
| Strain | MIC | MFC | MFC/MIC | MIC | MFC | MFC/MIC |
| C. albicans CBS 562 | 37.83 µM | 37.83 µM | 1 | 8.55 µM | 8.55 µM | 1 |
| C. albicans ATCC 90028 | 37.83 µM | 37.83 µM | 1 | 34.67 µM | 34.67 µM | 1 |
| C. krusei ATCC 6258 | 18.91 µM | 18.91 µM | 1 | 17.33 µM | 17.33 µM | 1 |
| C. tropicalis CBS 94 | 37.83 µM | 37.83 µM | 1 | 17.33 µM | 17.33 µM | 1 |
| C. glabrata ATCC 90030 | 18.91 µM | 18.91 µM | 1 | 8.55 µM | 8.55 µM | 1 |
| C. krusei CBS 573 | 18.91 µM | 18.91 µM | 1 | 34.67 µM | 34.67 µM | 1 |
| Cinnamaldehyde | Caspofungin | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Concentration (μM) | C. albicans ATCC 90028 | C. tropicalis CBS 94 | C. albicans ATCC 90028 | C. tropicalis CBS 94 | |||||
| Without Sorbitol | With Sorbitol | Without Sorbitol | With Sorbitol | Without Sorbitol | With Sorbitol | Without Sorbitol | With Sorbitol | ||
| 302.6 | − | − | − | − | 3.6 | − | − | − | − |
| 151.3 | − | − | − | − | 1.8 | − | − | − | − |
| 75.6 | − | − | − | − | 0.9 | − | − | − | − |
| 37.8 | − | − | − | − | 0.4 | − | − | − | − |
| 18.9 | + | + | + | + | 0.2 | − | − | − | − |
| 9.4 | + | + | + | + | 0.1 | − | − | − | − |
| 4.7 | + | + | + | + | 0.05 | − | + | − | + |
| 2.3 | + | + | + | + | 0.02 | + | + | + | + |
| Cinnamaldehyde | Nystatin | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Concentration (μM) | C. albicans ATCC 90028 | C. tropicalis CBS 94 | C. albicans ATCC90028 | C. tropicalis CBS 94 | |||||
| Without Ergosterol | With Ergosterol | Without Ergosterol | With Ergosterol | Without Ergosterol | With Ergosterol | Without Ergosterol | With Ergosterol | ||
| 302.6 | − | + | − | + | 129.5 | − | + | − | + |
| 151.3 | − | + | − | + | 64.7 | − | + | − | + |
| 75.6 | − | + | − | + | 32.3 | − | + | − | + |
| 37.8 | − | + | − | + | 16.1 | − | + | − | + |
| 18.9 | + | + | + | + | 8.0 | − | + | − | + |
| 9.4 | + | + | + | + | 4.0 | + | + | − | + |
| 4.7 | + | + | + | + | 1.9 | + | + | + | + |
| 2.3 | + | + | + | + | 1.0 | + | + | + | + |
| Sample | DPPH Method |
|---|---|
| IC50 (mM) | |
| Cinnamaldehyde | 1.74 |
| Trolox | 1.88 |
Sample Availability: Samples of the compounds are available from the authors. | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
da Nóbrega Alves, D.; Monteiro, A.F.M.; Andrade, P.N.; Lazarini, J.G.; Abílio, G.M.F.; Guerra, F.Q.S.; Scotti, M.T.; Scotti, L.; Rosalen, P.L.; Castro, R.D.d. Docking Prediction, Antifungal Activity, Anti-Biofilm Effects on Candida spp., and Toxicity against Human Cells of Cinnamaldehyde. Molecules 2020, 25, 5969. https://doi.org/10.3390/molecules25245969
da Nóbrega Alves D, Monteiro AFM, Andrade PN, Lazarini JG, Abílio GMF, Guerra FQS, Scotti MT, Scotti L, Rosalen PL, Castro RDd. Docking Prediction, Antifungal Activity, Anti-Biofilm Effects on Candida spp., and Toxicity against Human Cells of Cinnamaldehyde. Molecules. 2020; 25(24):5969. https://doi.org/10.3390/molecules25245969
Chicago/Turabian Styleda Nóbrega Alves, Danielle, Alex France Messias Monteiro, Patrícia Néris Andrade, Josy Goldoni Lazarini, Gisely Maria Freire Abílio, Felipe Queiroga Sarmento Guerra, Marcus Tullius Scotti, Luciana Scotti, Pedro Luiz Rosalen, and Ricardo Dias de Castro. 2020. "Docking Prediction, Antifungal Activity, Anti-Biofilm Effects on Candida spp., and Toxicity against Human Cells of Cinnamaldehyde" Molecules 25, no. 24: 5969. https://doi.org/10.3390/molecules25245969
APA Styleda Nóbrega Alves, D., Monteiro, A. F. M., Andrade, P. N., Lazarini, J. G., Abílio, G. M. F., Guerra, F. Q. S., Scotti, M. T., Scotti, L., Rosalen, P. L., & Castro, R. D. d. (2020). Docking Prediction, Antifungal Activity, Anti-Biofilm Effects on Candida spp., and Toxicity against Human Cells of Cinnamaldehyde. Molecules, 25(24), 5969. https://doi.org/10.3390/molecules25245969