Small Molecules Targeting the Specific Domains of Histone-Mark Readers in Cancer Therapy
Abstract
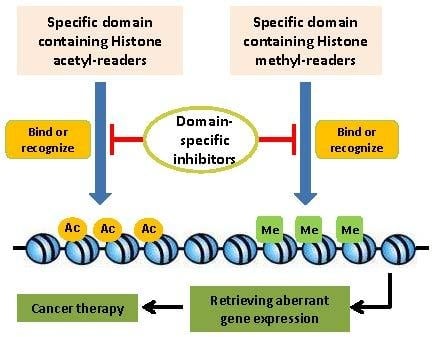
:1. Introduction
2. Proteins that Recognize Histone Modification Sites
2.1. Histone Acetylation Readers
2.1.1. BRD-Containing Proteins
2.1.2. PHD Finger Proteins
2.1.3. YEATS Proteins
2.2. Histone Methylation Readers
2.2.1. Chromodomain Proteins
2.2.2. PHD Finger Proteins
2.2.3. Tudor Domain Proteins
2.2.4. PWWP Motif Proteins
2.2.5. WDR and MBT Domain Proteins
3. Domain-Specific Inhibitors that Target Histone-Mark Readers
3.1. BET/BRD Inhibitors
3.2. PHD Inhibitors
3.3. BMT and Tudor Inhibitors
3.4. CHD Inhibitors
4. Conclusions and Perspectives
Author Contributions
Funding
Conflicts of Interest
Abbreviations
References
- Azad, G.K.; Tomar, R.S. Proteolytic clipping of histone tails: The emerging role of histone proteases in regulation of various biological processes. Mol. Biol. Rep. 2014, 41, 2717–2730. [Google Scholar] [CrossRef]
- Turner, B.M. Reading signals on the nucleosome with a new nomenclature for modified histones. Nat. Struct. Mol. Biol. 2005, 12, 110–112. [Google Scholar] [CrossRef]
- North, J.A.; Šimon, M.; Ferdinand, M.B.; Shoffner, M.A.; Picking, J.W.; Howard, C.J.; Mooney, A.M.; van Noort, J.; Poirier, M.G.; Ottesen, J.J. Histone H3 phosphorylation near the nucleosome dyad alters chromatin structure. Nucleic Acids. Res. 2014, 42, 4922–4933. [Google Scholar] [CrossRef] [Green Version]
- Li, G.; Zhu, P. Structure and organization of chromatin fiber in the nucleus. FEBS Lett. 2015, 589, 2893–2904. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ptashne, M. On the use of the word ‘epigenetic’. Curr. Biol. 2007, 17, R233–R236. [Google Scholar] [CrossRef] [Green Version]
- Moore, L.D.; Le, T.; Fan, G. DNA methylation and its basic function. Neuropsychopharmacology 2013, 38, 23–38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kornberg, R.D. Chromatin structure: A repeating unit of histones and DNA. Science 1974, 184, 868–871. [Google Scholar] [CrossRef] [PubMed]
- Kornberg, R.D.; Lorch, Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell 1999, 98, 285–294. [Google Scholar] [CrossRef] [Green Version]
- Bennett, R.L.; Licht, J.D. Targeting epigenetics in cancer. Annu. Rev. Pharmacol. Toxicol. 2018, 58, 187–207. [Google Scholar] [CrossRef]
- Yang, A.Y.; Kim, H.; Li, W.; Kong, A.N. Compound-derived epigenetic regulators targeting epigenetic readers, writers and erasers. Curr. Top. Med. Chem. 2016, 16, 697–713. [Google Scholar] [CrossRef]
- Mio, C.; Bulotta, S.; Russo, D.; Damante, G. Reading cancer: Chromatin readers as druggable targets for cancer treatment. Cancers 2019, 11, 61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shahbazian, M.D.; Grunstein, M. Functions of site-specific histone acetylation and deacetylation. Annu. Rev. Biochem. 2007, 76, 75–100. [Google Scholar] [CrossRef] [PubMed]
- Haberland, M.; Montgomery, R.L.; Olson, E.N. The many roles of histone deacetylases in development and physiology. Nat. Rev. Genet 2009, 10, 32–42. [Google Scholar] [CrossRef] [PubMed]
- Bannister, A.J.; Kouzarides, T. Regulation of chromatin by histone modifications. Cell Res. 2011, 21, 381–395. [Google Scholar] [CrossRef]
- Chen, H.; Tini, M.; Evans, R.M. HATS on and beyond chromatin. Curr. Opin. Cell Biol. 2001, 13, 218–224. [Google Scholar] [CrossRef]
- Biswas, S.; Rao, C.M. Epigenetic tools (The Writers, The Readers and The Erasers) and their implications in cancer therapy. Eur. J. Pharmacol. 2018, 837, 8–24. [Google Scholar] [CrossRef]
- Fujisawa, T.; Filippakopoulos, P. Functions of bromodomain-containing proteins and their roles in homeostasis and cancer. Nat. Rev. Mol. Cell Biol. 2017, 18, 246–262. [Google Scholar] [CrossRef]
- Filippakopoulos, P.; Picaud, S.; Mangos, M.; Keates, T.; Lambert, J.P.; Barsyte-Lovejoy, D.; Felletar, I.; Volkmer, R.; Müller, S.; Pawson, T.; et al. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell 2012, 149, 214–231. [Google Scholar] [CrossRef] [Green Version]
- Arrowsmith, C.H.; Bountra, C.; Fish, P.V.; Lee, K.; Schapira, M. Epigenetic protein families: A new frontier for drug discovery. Nat. Rev. Drug Discov. 2012, 11, 384–400. [Google Scholar] [CrossRef] [Green Version]
- Hudson, B.P.; Martinez-Yamout, M.A.; Dyson, H.J.; Wright, P.E. Solution structure and acetyl-lysine binding activity of the GCN5 bromodomain. J. Mol. Biol. 2000, 304, 355–370. [Google Scholar] [CrossRef]
- Dhalluin, C.; Carlson, J.E.; Zeng, L.; He, C.; Aggarwal, A.K.; Zhou, M.M. Structure and ligand of a histone acetyltransferase bromodomain. Nature 1999, 399, 491–496. [Google Scholar] [CrossRef] [PubMed]
- Zeng, L.; Zhang, Q.; Gerona-Navarro, G.; Moshkina, N.; Zhou, M.M. Structural basis of site-specific histone recognition by the bromodomains of human coactivators PCAF and CBP/p300. Structure 2008, 16, 643–652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakamura, T.; Mori, T.; Tada, S.; Krajewski, W.; Rozovskaia, T.; Wassell, R.; Dubois, G.; Mazo, A.; Croce, C.M.; Canaani, E. ALL-1 is a histone methyltransferase that assembles a supercomplex of proteins involved in transcriptional regulation. Mol. Cell 2002, 10, 1119–1128. [Google Scholar] [CrossRef]
- Zhu, L.; Li, Q.; Wong, S.H.; Huang, M.; Klein, B.J.; Shen, J.; Ikenouye, L.; Onishi, M.; Schneidawind, D.; Buechele, C.; et al. ASH1L Links Histone H3 Lysine 36 Dimethylation to MLL Leukemia. Cancer Discov. 2016, 6, 770–783. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, X.; Su, J.; Wang, F.; Liu, D.; Ding, J.; Yang, Y.; Conaway, J.W.; Conaway, R.C.; Cao, L.; Wu, D.; et al. Crosstalk between NSL histone acetyltransferase and MLL/SET complexes: NSL complex functions in promoting histone H3K4 di-methylation activity by MLL/SET complexes. PLoS Genet 2013, 9, e1003940. [Google Scholar] [CrossRef] [PubMed]
- Ewing, A.K.; Attner, M.; Chakravarti, D. Novel regulatory role for human Acf1 in transcriptional repression of vitamin D3 receptor-regulated genes. Mol. Endocrinol. 2007, 21, 1791–1806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruthenburg, A.J.; Li, H.; Milne, T.A.; Dewell, S.; McGinty, R.K.; Yuen, M.; Ueberheide, B.; Dou, Y.; Muir, T.W.; Patel, D.J.; et al. Recognition of a mononucleosomal histone modification pattern by BPTF via multivalent interactions. Cell 2011, 145, 692–706. [Google Scholar] [CrossRef] [Green Version]
- LeRoy, G.; Rickards, B.; Flint, S.J. The double bromodomain proteins Brd2 and Brd3 couple histone acetylation to transcription. Mol. Cell 2008, 30, 51–60. [Google Scholar] [CrossRef] [Green Version]
- Wu, T.; Kamikawa, Y.F.; Donohoe, M.E. Brd4’s bromodomains mediate histone H3 acetylation and chromatin remodeling in pluripotent cells through P300 and Brg1. Cell Rep. 2018, 25, 1756–1771. [Google Scholar] [CrossRef] [Green Version]
- Morinière, J.; Rousseaux, S.; Steuerwald, U.; Soler-López, M.; Curtet, S.; Vitte, A.L.; Govin, J.; Gaucher, J.; Sadoul, K.; Hart, D.J.; et al. Cooperative binding of two acetylation marks on a histone tail by a single bromodomain. Nature 2009, 461, 664–668. [Google Scholar]
- Fujiki, R.; Kim, M.S.; Sasaki, Y.; Yoshimura, K.; Kitagawa, H.; Kato, S. Ligand-induced transrepression by VDR through association of WSTF with acetylated histones. EMBO J. 2005, 24, 3881–3894. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Couture, J.P.; Nolet, G.; Beaulieu, E.; Blouin, R.; Gévry, N. The p400/Brd8 chromatin remodeling complex promotes adipogenesis by incorporating histone variant H2A.Z at PPARγ target genes. Endocrinology 2012, 153, 5796–5808. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalla, C.; Nentwich, H.; Schlotter, M.; Mertens, D.; Wildenberger, K.; Döhner, H.; Stilgenbauer, S.; Lichter, P. Translocation t(X;11)(q13;q23) in B-cell chronic lymphocytic leukemia disrupts two novel genes. Genes Chromosomes Cancer 2005, 42, 128–143. [Google Scholar] [CrossRef] [PubMed]
- Tropberger, P.; Pott, S.; Keller, C.; Kamieniarz-Gdula, K.; Caron, M.; Richter, F.; Li, G.; Mittler, G.; Liu, E.T.; Bühler, M.; et al. Regulation of transcription through acetylation of H3K122 on the lateral surface of the histone octamer. Cell 2013, 152, 859–872. [Google Scholar] [CrossRef] [Green Version]
- Jin, Q.; Yu, L.R.; Wang, L.; Zhang, Z.; Kasper, L.H.; Lee, J.E.; Wang, C.; Brindle, P.K.; Dent, S.Y.; Ge, K. Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J. 2011, 30, 249–262. [Google Scholar] [CrossRef] [PubMed]
- Podcheko, A.; Northcott, P.; Bikopoulos, G.; Lee, A.; Bommareddi, S.R.; Kushner, J.A.; Farhang-Fallah, J.; Rozakis-Adcock, M. Identification of a WD40 repeat-containing isoform of PHIP as a novel regulator of beta-cell growth and survival. Mol. Cell Biol. 2007, 27, 6484–6496. [Google Scholar] [CrossRef] [Green Version]
- Huang, H.; Rambaldi, I.; Daniels, E.; Featherstone, M. Expression of the Wdr9 gene and protein products during mouse development. Dev. Dyn. 2003, 227, 608–614. [Google Scholar] [CrossRef]
- Koo, S.J.; Fernandez-Montalvan, A.E.; Badock, V.; Ott, C.J.; Holton, S.J.; von Ahsen, O.; Toedling, J.; Vittori, S.; Bradner, J.E.; Gorjanacz, M. ATAD2 is an epigenetic reader of newly synthesized histone marks during DNA replication. Oncotarget 2016, 7, 70323–70335. [Google Scholar] [CrossRef] [Green Version]
- Revenko, A.S.; Kalashnikova, E.V.; Gemo, A.T.; Zou, J.X.; Chen, H.W. Chromatin loading of E2F-MLL complex by cancer-associated coregulator ANCCA via reading a specific histone mark. Mol. Cell Biol. 2010, 30, 5260–5272. [Google Scholar] [CrossRef] [Green Version]
- Mishima, Y.; Miyagi, S.; Saraya, A.; Negishi, M.; Endoh, M.; Endo, T.A.; Toyoda, T.; Shinga, J.; Katsumoto, T.; Chiba, T.; et al. The Hbo1-Brd1/Brpf2 complex is responsible for global acetylation of H3K14 and required for fetal liver erythropoiesis. Blood 2011, 118, 2443–2453. [Google Scholar] [CrossRef]
- Sun, H.; Liu, J.; Zhang, J.; Shen, W.; Huang, H.; Xu, C.; Dai, H.; Wu, J.; Shi, Y. Solution structure of BRD7 bromodomain and its interaction with acetylated peptides from histone H3 and H4. Biochem. Biophys. Res. Commun. 2007, 358, 435–441. [Google Scholar] [CrossRef] [PubMed]
- Flynn, E.M.; Huang, O.W.; Poy, F.; Oppikofer, M.; Bellon, S.F.; Tang, Y.; Cochran, A.G. A subset of human bromodomains recognizes butyryllysine and crotonyllysine histone peptide modifications. Structure 2015, 23, 1801–1814. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poplawski, A.; Hu, K.; Lee, W.; Natesan, S.; Peng, D.; Carlson, S.; Shi, X.; Balaz, S.; Markley, J.L.; Glass, K.C. Molecular insights into the recognition of N-terminal histone modifications by the BRPF1 bromodomain. J. Mol. Biol. 2014, 426, 1661–1676. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Vlassis, A.; Roques, C.; Lalonde, M.E.; González-Aguilera, C.; Lambert, J.P.; Lee, S.B.; Zhao, X.; Alabert, C.; et al. BRPF3-HBO1 regulates replication origin activation and histone H3K14 acetylation. EMBO J. 2016, 35, 176–192. [Google Scholar] [CrossRef]
- Zhou, Y.; Grummt, I. The PHD finger/bromodomain of NoRC interacts with acetylated histone H4K16 and is sufficient for rDNA silencing. Curr. Biol. 2005, 15, 1434–1438. [Google Scholar] [CrossRef] [Green Version]
- Ferguson, F.M.; Dias, D.M.; Rodrigues, J.P.; Wienk, H.; Boelens, R.; Bonvin, A.M.; Abell, C.; Ciulli, A. Binding hotspots of BAZ2B bromodomain: Histone interaction revealed by solution NMR driven docking. Biochemistry 2014, 53, 6706–6716. [Google Scholar] [CrossRef] [Green Version]
- Seeler, J.S.; Marchio, A.; Sitterlin, D.; Transy, C.; Dejean, A. Interaction of SP100 with HP1 proteins: A link between the promyelocytic leukemia-associated nuclear bodies and the chromatin compartment. Proc. Natl. Acad. Sci. USA 1998, 95, 7316–7321. [Google Scholar] [CrossRef] [Green Version]
- Bloch, D.B.; Nakajima, A.; Gulick, T.; Chiche, J.D.; Orth, D.; de La Monte, S.M.; Bloch, K.D. Sp110 localizes to the PML-Sp100 nuclear body and may function as a nuclear hormone receptor transcriptional coactivator. Mol. Cell Biol. 2000, 20, 6138–6146. [Google Scholar] [CrossRef] [Green Version]
- Karaky, M.; Fedetz, M.; Potenciano, V.; Andrés-León, E.; Codina, A.E.; Barrionuevo, C.; Alcina, A.; Matesanz, F. SP140 regulates the expression of immune-related genes associated with multiple sclerosis and other autoimmune diseases by NF-κB inhibition. Hum. Mol. Genet 2018, 27, 4012–4023. [Google Scholar] [CrossRef]
- Franke, A.; McGovern, D.P.; Barrett, J.C.; Wang, K.; Radford-Smith, G.L.; Ahmad, T.; Lees, C.W.; Balschun, T.; Lee, J.; et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat. Genet 2010, 42, 1118–1125. [Google Scholar] [CrossRef] [Green Version]
- Lv, D.; Li, Y.; Zhang, W.; Alvarez, A.A.; Song, L.; Tang, J.; Gao, W.Q.; Hu, B.; Cheng, S.Y.; Feng, H. TRIM24 is an oncogenic transcriptional co-activator of STAT3 in glioblastoma. Nat. Commun. 2017, 8, 1454. [Google Scholar] [CrossRef] [PubMed]
- Agricola, E.; Randall, R.A.; Gaarenstroom, T.; Dupont, S.; Hill, C.S. Recruitment of TIF1gamma to chromatin via its PHD finger-bromodomain activates its ubiquitin ligase and transcriptional repressor activities. Mol. Cell 2011, 43, 85–96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khetchoumian, K.; Teletin, M.; Mark, M.; Lerouge, T.; Cerviño, M.; Oulad-Abdelghani, M.; Chambon, P.; Losson, R. TIF1delta, a novel HP1-interacting member of the transcriptional intermediary factor 1 (TIF1) family expressed by elongating spermatids. J. Biol. Chem. 2004, 279, 48329–48341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jacobson, R.H.; Ladurner, A.G.; King, D.S.; Tjian, R. Structure and function of a human TAFII250 double bromodomain module. Science 2000, 288, 1422–1425. [Google Scholar] [CrossRef]
- Wang, P.J.; Page, D.C. Functional substitution for TAF(II)250 by a retroposed homolog that is expressed in human spermatogenesis. Hum. Mol. Genet 2002, 11, 2341–2346. [Google Scholar] [CrossRef] [Green Version]
- Savitsky, P.; Krojer, T.; Fujisawa, T.; Lambert, J.P.; Picaud, S.; Wang, C.Y.; Shanle, E.K.; Krajewski, K.; Friedrichsen, H.; Kanapin, A.; et al. Multivalent Histone and DNA engagement by a PHD/BRD/PWWP triple reader cassette recruits ZMYND8 to K14ac-rich chromatin. Cell Rep. 2016, 17, 2724–2737. [Google Scholar] [CrossRef] [Green Version]
- Wei, G.; Schaffner, A.E.; Baker, K.M.; Mansky, K.C.; Ostrowski, M.C. Ets-2 interacts with co-repressor BS69 to repress target gene expression. Anticancer Res. 2003, 23, 2173–2178. [Google Scholar]
- Chandrasekaran, R.; Thompson, M. Polybromo-1-bromodomains bind histone H3 at specific acetyl-lysine positions. Biochem. Biophys. Res. Commun. 2007, 355, 661–666. [Google Scholar] [CrossRef]
- Morrison, E.A.; Sanchez, J.C.; Ronan, J.L.; Farrell, D.P.; Varzavand, K.; Johnson, J.K.; Gu, B.X.p; Crabtree, G.R.; Musselman, C.A. DNA binding drives the association of BRG1/hBRM bromodomains with nucleosomes. Nat. Commun. 2017, 8, 16080. [Google Scholar] [CrossRef]
- Pham, T.X.; Bae, M.; Lee, Y.; Park, Y.K.; Lee, J.Y. Transcriptional and posttranscriptional repression of histone deacetylases by docosahexaenoic acid in macrophages. J. Nutr. Biochem. 2018, 57, 162–169. [Google Scholar] [CrossRef]
- Attar, N.; Kurdistani, S.K. Exploitation of EP300 and CREBBP lysine acetyltransferases by cancer. Cold Spring Harb. Perspect. Med. 2017, 7, a026534. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Musco, G.; Peterson, P. PHD finger of autoimmune regulator: An epigenetic link between the histone modifications and tissue-specific antigen expression in thymus. Epigenetics 2008, 3, 310–314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Jang, Y.; Lee, J.E.; Ahn, J.; Xu, L.; Holden, M.R.; Cornett, E.M.; Krajewski, K.; Klein, B.J.; Wang, S.P.; et al. Selective binding of the PHD6 finger of MLL4 to histone H4K16ac links MLL4 and MOF. Nat. Commun. 2019, 10, 2314. [Google Scholar] [CrossRef] [PubMed]
- Sabari, B.R.; Zhang, D.; Allis, C.D.; Zhao, Y. Metabolic regulation of gene expression through histone acylations. Nat. Rev. Mol. Cell Biol. 2017, 18, 90–101. [Google Scholar] [CrossRef] [Green Version]
- Qiu, Y.; Liu, L.; Zhao, C.; Han, C.; Li, F.; Zhang, J.; Wang, Y.; Li, G.; Mei, Y.; Wu, M.; et al. Combinatorial readout of unmodified H3R2 and acetylated H3K14 by the tandem PHD finger of MOZ reveals a regulatory mechanism for HOXA9 transcription. Genes Dev. 2012, 26, 1376–1391. [Google Scholar] [CrossRef] [Green Version]
- Ali, M.; Yan, K.; Lalonde, M.E.; Degerny, C.; Rothbart, S.B.; Strahl, B.D.; Côté, J.; Yang, X.J.; Kutateladze, T.G. Tandem PHD fingers of MORF/MOZ acetyltransferases display selectivity for acetylated histone H3 and are required for the association with chromatin. J. Mol. Biol. 2012, 424, 328–338. [Google Scholar] [CrossRef] [Green Version]
- Huber, F.M.; Greenblatt, S.M.; Davenport, A.M.; Martinez, C.; Xu, Y.; Vu, L.P.; Nimer, S.D.; Hoelz, A. Histone-binding of DPF2 mediates its repressive role in myeloid differentiation. Proc. Natl. Acad. Sci. USA 2017, 114, 6016–6021. [Google Scholar] [CrossRef] [Green Version]
- Lange, M.; Kaynak, B.; Forster, U.B.; Tönjes, M.; Fischer, J.J.; Grimm, C.; Schlesinger, J.; Just, S.; Dunkel, I.; Krueger, T.; et al. Regulation of muscle development by DPF3, a novel histone acetylation and methylation reader of the BAF chromatin remodeling complex. Genes Dev. 2008, 22, 2370–2384. [Google Scholar] [CrossRef] [Green Version]
- Le, M.; Yu, D.Y.; Jensen, K.; Chevalier, A.; Courbeyrette, R.; Boulard, Y.; Smith, M.M.; Mann, C. Yaf9, a novel NuA4 histone acetyltransferase subunit, is required for the cellular response to spindle stress in yeast. Mol. Cell Biol. 2003, 23, 6086–6102. [Google Scholar]
- Schulze, J.M.; Wang, A.Y.; Kobor, M.S. YEATS domain proteins: A diverse family with many links to chromatin modification and transcription. Biochem. Cell Biol. 2009, 87, 65–75. [Google Scholar] [CrossRef]
- Daser, A.; Rabbitts, T.H. Extending the repertoire of the mixed-lineage leukemia gene MLL in leukemogenesis. Genes Dev. 2004, 18, 965–974. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Wen, H.; Xi, Y.; Tanaka, K.; Wang, H.; Peng, D.; Ren, Y.; Jin, Q.; Dent, S.Y.; Li, W.; et al. AF9 YEATS domain links histone acetylation to DOT1L-mediated H3K79 methylation. Cell 2014, 159, 558–571. [Google Scholar] [CrossRef] [Green Version]
- Wan, L.; Wen, H.; Li, Y.; Lyu, J.; Xi, Y.; Hoshii, T.; Joseph, J.K.; Wang, X.; Loh, Y.E.; Erb, M.A.; et al. ENL links histone acetylation to oncogenic gene expression in acute myeloid leukaemia. Nature 2017, 543, 265–269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cho, H.J.; Li, H.; Linhares, B.M.; Kim, E.; Ndoj, J.; Miao, H.; Grembecka, J.; Cierpicki, T. GAS41 recognizes di-acetylated histone H3 through a bivalent binding mode. ACS Chem. Biol. 2018, 13, 2739–2746. [Google Scholar] [CrossRef]
- Shanle, E.K.; Andrews, F.H.; Meriesh, H.; McDaniel, S.L.; Dronamraju, R.; DiFiore, J.V.; Jha, D.; Wozniak, G.G.; Bridgers, J.B.; Kerschner, J.L.; et al. Association of Taf14 with acetylated histone H3 directs gene transcription and the DNA damage response. Genes Dev. 2015, 29, 1795–1800. [Google Scholar] [CrossRef] [Green Version]
- Wang, A.Y.; Schulze, J.M.; Skordalakes, E.; Gin, J.W.; Berger, J.M.; Rine, J.; Kobor, M.S. Asf1-like structure of the conserved Yaf9 YEATS domain and role in H2A.Z deposition and acetylation. Proc. Natl. Acad. Sci. USA 2009, 106, 21573–21578. [Google Scholar] [CrossRef] [Green Version]
- Musselman, C.A.; Lalonde, M.E.; Côté, J.; Kutateladze, T.G. Perceiving the epigenetic landscape through histone readers. Nat. Struct. Mol. Biol. 2012, 19, 1218–1227. [Google Scholar] [CrossRef] [Green Version]
- Ball, L.J.; Murzina, N.V.; Broadhurst, R.W.; Raine, A.R.; Archer, S.J.; Stott, F.J.; Murzin, A.G.; Singh, P.B.; Domaille, P.J.; Laue, E.D. Structure of the chromatin binding (chromo) domain from mouse modifier protein 1. EMBO J. 1997, 16, 2473–2481. [Google Scholar] [CrossRef] [Green Version]
- Yap, K.L.; Zhou, M.M. Keeping it in the family: Diverse histone recognition by conserved structural folds. Crit. Rev. Biochem. Mol. Biol. 2010, 45, 488–505. [Google Scholar] [CrossRef] [Green Version]
- Eissenberg, J.C. Structural biology of the chromodomain: Form and function. Gene 2012, 496, 69–78. [Google Scholar] [CrossRef]
- Kaustov, L.; Ouyang, H.; Amaya, M.; Lemak, A.; Nady, N.; Duan, S.; Wasney, G.A.; Li, Z.; Vedadi, M.; Schapira, M.; et al. Recognition and specificity determinants of the human Cbx chromodomain. J. Biol. Chem. 2011, 286, 521–529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schalch, T.; Job, G.; Noffsinger, V.J.; Shanker, S.; Kuscu, C.; Joshua-Tor, L.; Partridge, J.F. High-affinity binding of Chp1 chromodomain to K9 methylated histone H3 is required to establish centromeric heterochromatin. Mol. Cell 2009, 34, 36–46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sadaie, M.; Kawaguchi, R.; Ohtani, Y.; Arisaka, F.; Tanaka, K.; Shirahige, K.; Nakayama, J. Balance between distinct HP1 family proteins controls heterochromatin assembly in fission yeast. Mol. Cell Biol. 2008, 28, 6973–6988. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vermeulen, M.; Eberl, H.C.; Matarese, F.; Marks, H.; Denissov, S.; Butter, F.; Lee, K.K.; Olsen, J.V.; Hyman, A.; Stunnenberg, H.G.; et al. Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers. Cell 2010, 142, 967–980. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bernstein, E.; Duncan, E.M.; Masui, O.; Gil, J.; Heard, E.; Allis, C.D. Mouse polycomb proteins bind differentially to methylated histone H3 and RNA and are enriched in facultative heterochromatin. Mol. Cell Biol. 2006, 26, 2560–2569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Larschan, E.; Alekseyenko, A.A.; Gortchakov, A.A.; Peng, S.; Li, B.; Yang, P.; Workman, J.L.; Park, P.J.; Kuroda, M.I. MSL complex is attracted to genes marked by H3K36 trimethylation using a sequence-independent mechanism. Mol. Cell 2007, 28, 121–133. [Google Scholar] [CrossRef]
- Kim, D.; Blus, B.J.; Chandra, V.; Huang, P.; Rastinejad, F.; Khorasanizadeh, S. Corecognition of DNA and a methylated histone tail by the MSL3 chromodomain. Nat. Struct. Mol. Biol. 2010, 17, 1027–1029. [Google Scholar] [CrossRef] [Green Version]
- Zhang, P.; Du, J.; Sun, B.; Dong, X.; Xu, G.; Zhou, J.; Huang, Q.; Liu, Q.; Hao, Q.; Ding, J. Structure of human MRG15 chromo domain and its binding to Lys36-methylated histone H3. Nucl. Acids. Res. 2006, 34, 6621–6628. [Google Scholar] [CrossRef]
- Joshi, A.A.; Struhl, K. Eaf3 chromodomain interaction with methylated H3-K36 links histone deacetylation to Pol II elongation. Mol. Cell 2005, 20, 971–978. [Google Scholar] [CrossRef]
- Flanagan, J.F.; Mi, L.Z.; Chruszcz, M.; Cymborowski, M.; Clines, K.L.; Kim, Y.; Minor, W.; Rastinejad, F.; Khorasanizadeh, S. Double chromodomains cooperate to recognize the methylated histone H3 tail. Nature 2005, 438, 1181–1185. [Google Scholar] [CrossRef]
- Sims, R.J., 3rd; Millhouse, S.; Chen, C.F.; Lewis, B.A.; Erdjument-Bromage, H.; Tempst, P.; Manley, J.L.; Reinberg, D. Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Mol. Cell 2007, 28, 665–676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sims, R.J., 3rd; Chen, C.F.; Santos-Rosa, H.; Kouzarides, T.; Patel, S.S.; Reinberg, D. Human but not yeast CHD1 binds directly and selectively to histone H3 methylated at lysine 4 via its tandem chromodomains. J. Biol. Chem. 2005, 280, 41789–41792. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schnetz, M.P.; Bartels, C.F.; Shastri, K.; Balasubramanian, D.; Zentner, G.E.; Balaji, R.; Zhang, X.; Song, L.; Wang, Z.; Laframboise, T.; et al. Genomic distribution of CHD7 on chromatin tracks H3K4 methylation patterns. Genome Res. 2009, 19, 590–601. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.; Daniel, J.; Espejo, A.; Lake, A.; Krishna, M.; Xia, L.; Zhang, Y.; Bedford, M.T. Tudor, MBT and chromo domains gauge the degree of lysine methylation. EMBO Rep. 2006, 7, 397–403. [Google Scholar] [CrossRef]
- Machev, N.; Saut, N.; Longepied, G.; Terriou, P.; Navarro, A.; Levy, N.; Guichaoua, M.; Metzler-Guillemain, C.; Collignon, P.; Frances, A.M.; et al. Sequence family variant loss from the AZFc interval of the human Y chromosome, but not gene copy loss, is strongly associated with male infertility. J. Med. Genet 2004, 41, 814–825. [Google Scholar] [CrossRef] [Green Version]
- Franz, H.; Mosch, K.; Soeroes, S.; Urlaub, H.; Fischle, W. Multimerization and H3K9me3 binding are required for CDYL1b heterochromatin association. J. Biol. Chem. 2009, 284, 35049–35059. [Google Scholar] [CrossRef] [Green Version]
- Escamilla-Del-Arenal, M.; da Rocha, S.T.; Spruijt, C.G.; Masui, O.; Renaud, O.; Smits, A.H.; Margueron, R.; Vermeulen, M.; Heard, E. Cdyl, a new partner of the inactive X chromosome and potential reader of H3K27me3 and H3K9me2. Mol. Cell Biol. 2013, 33, 5005–5020. [Google Scholar] [CrossRef] [Green Version]
- Vermeulen, M.; Mulder, K.W.; Denissov, S.; Pijnappel, W.W.; van Schaik, F.M.; Varier, R.A.; Baltissen, M.P.; Stunnenberg, H.G.; Mann, M.; Timmers, H.T. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell 2007, 131, 58–69. [Google Scholar] [CrossRef] [Green Version]
- Klein, B.J.; Wang, X.; Cui, G.; Yuan, C.; Botuyan, M.V.; Lin, K.; Lu, Y.; Wang, X.; Zhao, Y.; Bruns, C.J.; et al. PHF20 readers link methylation of histone H3K4 and p53 with H4K16 acetylation. Cell Rep. 2016, 17, 1158–1170. [Google Scholar] [CrossRef] [Green Version]
- Ali, M.; Rincón-Arano, H.; Zhao, W.; Rothbart, S.B.; Tong, Q.; Parkhurst, S.M.; Strahl, B.D.; Deng, L.W.; Groudine, M.; Kutateladze, T.G. Molecular basis for chromatin binding and regulation of MLL5. Proc. Natl. Acad. Sci. USA 2013, 110, 11296–11301. [Google Scholar] [CrossRef] [Green Version]
- Xie, S.; Jakoncic, J.; Qian, C. UHRF1 double Tudor domain and the adjacent PHD finger act together to recognize K9me3-containing histone H3 tail. J. Mol. Biol. 2012, 415, 318–328. [Google Scholar] [CrossRef] [PubMed]
- Mansfield, R.E.; Musselman, C.A.; Kwan, A.H.; Oliver, S.S.; Garske, A.L.; Davrazou, F.; Denu, J.M.; Kutateladze, T.G.; Mackay, J.P. Plant homeodomain (PHD) fingers of CHD4 are histone H3-binding modules with preference for unmodified H3K4 and methylated H3K9. J. Biol. Chem. 2011, 286, 1779–11791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horton, J.R.; Upadhyay, A.K.; Qi, H.H.; Zhang, X.; Shi, Y.; Cheng, X. Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases. Nat. Struct. Mol. Biol. 2010, 17, 38–43. [Google Scholar] [CrossRef] [PubMed]
- Pek, J.W.; Anand, A.; Kai, T. Tudor domain proteins in development. Development 2012, 139, 2255–2266. [Google Scholar] [CrossRef] [Green Version]
- Musselman, C.A.; Avvakumov, N.; Watanabe, R.; Abraham, CG.; Lalonde, M.E.; Hong, Z.; Allen, C.; Roy, S.; Nuñez, J.K.; Nickoloff, J.; et al. Molecular basis for H3K36me3 recognition by the Tudor domain of PHF1. Nat. Struct. Mol. Biol. 2012, 19, 1266–1272. [Google Scholar] [CrossRef]
- Ballaré, C.; Lange, M.; Lapinaite, A.; Martin, GM.; Morey, L.; Pascual, G.; Liefke, R.; Simon, B.; Shi, Y.; Gozani, O.; et al. Phf19 links methylated Lys36 of histone H3 to regulation of Polycomb activity. Nat. Struct. Mol. Biol. 2012, 19, 1257–1265. [Google Scholar] [CrossRef] [Green Version]
- Cai, L.; Rothbart, S.B.; Lu, R.; Xu, B.; Chen, W.Y.; Tripathy, A.; Rockowitz, S.; Zheng, D.; Patel, D.J.; Allis, C.D.; et al. An H3K36 methylation-engaging Tudor motif of polycomb-like proteins mediates PRC2 complex targeting. Mol. Cell 2013, 49, 571–582. [Google Scholar] [CrossRef] [Green Version]
- Weaver, T.M.; Morrison, E.A.; Musselman, C.A. Reading more than histones: The prevalence of nucleic acid binding among reader domains. Molecules 2018, 23, 2614. [Google Scholar] [CrossRef] [Green Version]
- Shi, Y.; Whetstine, J.R. Dynamic Regulation of Histone Lysine Methylation by Demethylases. Mol. Cell 2007, 25, 1–14. [Google Scholar] [CrossRef]
- Lee, J.; Thompson, J.R.; Botuyan, M.V.; Mer, G. Distinct binding modes specify the recognition of methylated histones H3K4 and H4K20 by JMJD2A-tudor. Nat. Struct. Mol. Biol. 2008, 15, 109–111. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.; Fang, J.; Bedford, M.T.; Zhang, Y.; Xu, R.M. Recognition of histone H3 lysine-4 methylation by the double tudor domain of JMJD2A. Science 2006, 312, 748–751. [Google Scholar] [CrossRef]
- Lu, R.; Wang, G.G. Tudor: A versatile family of histone methylation ‘readers’. Trends Biochem. Sci. 2013, 38, 546–555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Botuyan, M.V.; Lee, J.; Ward, I.M.; Kim, J.E.; Thompson, J.R.; Chen, J.; Mer, G. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell 2006, 127, 1361–1373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, H.; Zeng, H.; Lam, R.; Tempel, W.; Amaya, M.F.; Xu, C.; Dombrovski, L.; Qiu, W.; Wang, Y.; Min, J. Structural and histone binding ability characterizations of human PWWP domains. PLoS ONE 2011, 6, e18919. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, R.; Gao, J.; Zhang, J.; Zhang, X.; Xu, C.; Liao, S.; Tu, X. Solution structure of TbTFIIS2-2 PWWP domain from Trypanosoma brucei and its binding to H4K17me3 and H3K32me3. Biochem. J. 2019, 476, 421–431. [Google Scholar] [CrossRef]
- Pradeepa, M.M.; Sutherland, H.G.; Ule, J.; Grimes, G.R.; Bickmore, W.A. Psip1/Ledgf p52 binds methylated histone H3K36 and splicing factors and contributes to the regulation of alternative splicing. PLoS Genet 2012, 8, e1002717. [Google Scholar] [CrossRef] [Green Version]
- Guo, R.; Zheng, L.J.; Park, J.W.; Lv, R.; Chen, H.; Jiao, F.; Xu, W.; Mu, S.; Wen, H.; Qiu, J.; et al. BS69/ZMYND11 reads and connects histone H3.3 lysine 36 trimethylation-decorated chromatin to regulated pre-mRNA processing. Mol. Cell 2014, 56, 298–310. [Google Scholar] [CrossRef] [Green Version]
- Cermáková, K.; Tesina, P.; Demeulemeester, J.; EI, A.S.; Méreau, H.; Schwaller, J.; Rezáčová, P.; Veverka, V.; De Rijck, J. Validation and structural characterization of the LEDGF/p75-MLL interface as a new target for the treatment of MLL-dependent leukemia. Cancer Res. 2014, 74, 5139–5151. [Google Scholar]
- Tesina, P.; Čermáková, K.; Hořejší, M.; Procházková, K.; Fábry, M.; Sharma, S.; Christ, F.; Demeulemeester, J.; Debyser, Z.; Rijck, J.; et al. Multiple cellular proteins interact with LEDGF/p75 through a conserved unstructured consensus motif. Nat. Commun. 2015, 6, 7968. [Google Scholar] [CrossRef] [Green Version]
- Sharma, S.; Čermáková, K.; De, R.J.; Demeulemeester, J.; Fábry, M.; El, A.S.; Van, B.S.; Lepšík, M.; Tesina, P.; Duchoslav, V.; et al. Affinity switching of the LEDGF/p75 IBD interactome is governed by kinase-dependent phosphorylation. Proc. Natl. Acad. Sci. USA 2018, 115, 7053–7062. [Google Scholar] [CrossRef] [Green Version]
- Gilbert, T.M.; McDaniel, S.L.; Byrum, S.D.; Cades, J.A.; Dancy, B.C.; Wade, H.; Tackett, A.J.; Strahl, B.D.; Taverna, S.D. A PWWP domain-containing protein targets the NuA3 acetyltransferase complex via histone H3 lysine 36 trimethylation to coordinate transcriptional elongation at coding regions. Mol. Cell Proteomics 2014, 13, 2883–2895. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Reddy, B.; Thompson, J.; Wang, H.; Noma, K.; Yates, J.R., 3rd; Jia, S. Regulation of Set9-mediated H4K20 methylation by a PWWP domain protein. Mol. Cell 2009, 33, 428–437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sendžikaitė, G.; Hanna, C.W.; Stewart-Morgan, K.R.; Ivanova, E.; Kelsey, G. A DNMT3A PWWP mutation leads to methylation of bivalent chromatin and growth retardation in mice. Nat. Commun. 2019, 10, 1884. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Y.; Zhang, W.; Zhao, C.; Wang, Y.; Wang, W.; Zhang, J.; Zhang, Z.; Li, G.; Shi, Y.; Tu, X.; et al. Solution structure of the Pdp1 PWWP domain reveals its unique binding sites for methylated H4K20 and DNA. Biochem. J. 2012, 442, 527–538. [Google Scholar] [CrossRef] [Green Version]
- Tian, W.; Yan, P.; Xu, N.; Chakravorty, A.; Liefke, R.; Xi, Q.; Wang, Z. The HRP3 PWWP domain recognizes the minor groove of double-stranded DNA and recruits HRP3 to chromatin. Nucl. Acids. Res. 2019, 47, 5436–5448. [Google Scholar] [CrossRef]
- Schapira, M.; Tyers, M.; Torrent, M.; Arrowsmith, C.H. WD-repeat domain proteins: A novel target class. Nat. Rev. Drug Discov. 2017, 16, 773–786. [Google Scholar] [CrossRef]
- Margueron, R.; Justin, N.; Ohno, K.; Sharpe, M.L.; Son, J.; Drury, W.J., 3rd. Role of the polycomb protein Eed in the propagation of repressive histone marks. Nature 2009, 461, 762–767. [Google Scholar] [CrossRef] [Green Version]
- Wysocka, J.; Swigut, T.; Milne, T.A.; Dou, Y.; Zhang, X.; Burlingame, A.L.; Roeder, R.G.; Brivanlou, A.H.; Allis, C.D. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell 2005, 121, 859–872. [Google Scholar] [CrossRef] [Green Version]
- Ruthenburg, A.J.; Wang, W.; Graybosch, D.M.; Li, H.; Allis, C.D.; Patel, D.J.; Verdine, G.L. Histone H3 recognition and presentation by the WDR5 module of the MLL1 complex. Nat. Struct. Mol. Biol. 2006, 13, 704–712. [Google Scholar] [CrossRef]
- Bonasio, R.; Lecona, E.; Reinberg, D. MBT domain proteins in development and disease. Semin. Cell Dev. Biol. 2010, 21, 221–230. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Fischle, W.; Wang, W.; Duncan, E.M.; Liang, L.; Murakami-Ishibe, S.; Allis, C.D.; Patel, D.J. Structural basis for lower lysine methylation state-specific readout by MBT repeats of L3MBTL1 and an engineered PHD finger. Mol. Cell 2007, 28, 677–691. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klymenko, T.; Papp, B.; Fischle, W.; Köcher, T.; Schelder, M.; Fritsch, C.; Wild, B.; Wilm, M.; Müller, J. A polycomb group protein complex with sequence-specific DNA-binding and selective methyl-lysine-binding activities. Genes Dev. 2006, 20, 1110–1122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, M.; Shen, H.; Jin, Y.; Lin, T.; Cai, Q.; Pinard, M.A.; Biswas, S.; Tran, Q.; Li, G.; Shenoy, A.K.; et al. The malignant brain tumor (MBT) domain protein SFMBT1 is an integral histone reader subunit of the LSD1 demethylase complex for chromatin association and epithelial-to-mesenchymal transition. J. Biol. Chem. 2013, 288, 27680–27691. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koester-Eiserfunke, N.; Fischle, W. H3K9me2/3 binding of the MBT domain protein LIN-61 is essential for caenorhabditis elegans vulva development. PLoS Genet 2011, 7, e1002017. [Google Scholar] [CrossRef] [Green Version]
- Waldmann, T.; Schneider, R. Targeting histone modifications-epigenetics in cancer. Curr. Opin. Cell Biol. 2013, 25, 184–189. [Google Scholar] [CrossRef]
- Cermakova, K.; Hodges, H.C. Next-generation drugs and probes for chromatin biology: From targeted protein degradation to phase separation. Molecules 2018, 23, 1958. [Google Scholar] [CrossRef] [Green Version]
- Romero, F.A.; Taylor, A.M.; Crawford, T.D.; Tsui, V.; Côté, A.; Magnuson, S. Disrupting acetyl-lysine recognition: Progress in the development of bromodomain inhibitors. J. Med. Chem. 2016, 59, 1271–1298. [Google Scholar] [CrossRef]
- Prinjha, R.K.; Witherington, J.; Lee, K. Place your BETs: The therapeutic potential of bromodomains. Trends Pharmacol. Sci. 2012, 33, 146–153. [Google Scholar] [CrossRef]
- Delmore, J.E.; Issa, G.C.; Lemieux, M.E.; Rahl, P.B.; Shi, J.; Jacobs, H.M.; Kastritis, E.; Gilpatrick, T.; Paranal, RM.; Qi, J.; et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 2011, 146, 904–917. [Google Scholar] [CrossRef] [Green Version]
- Zuber, J.; Shi, J.; Wang, E.; Rappaport, AR.; Herrmann, H.; Sison, E.A.; Magoon, D.; Qi, J.; Blatt, K.; Wunderlich, M.; et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature 2011, 478, 524–528. [Google Scholar] [CrossRef] [Green Version]
- Puissant, A.; Frumm, S.M.; Alexe, G.; Bassil, C.F.; Qi, J.; Chanthery, Y.H.; Nekritz, EA.; Zeid, R.; Gustafson, W.C.; Greninger, P.; et al. Targeting MYCN in neuroblastoma by BET bromodomain inhibition. Cancer Discov. 2013, 3, 308–323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lockwood, W.W.; Zejnullahu, K.; Bradner, J.E.; Varmus, H. Sensitivity of human lung adenocar- cinoma cell lines to targeted inhibition of BET epigenetic signaling proteins. Proc. Natl. Acad. Sci. USA 2012, 109, 19408–19413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia, P.L.; Miller, A.L.; Kreitzburg, K.M.; Council, L.N.; Gamblin, T.L.; Christei, J.D.; Heslin, M.J.; Arnoletti, J.P.; Richardson, J.H.; Chen, D.; et al. The BET bromodomain inhibitor JQ1 suppresses growth of pancreatic ductal adenocarcinoma in patient-derived xenograft models. Oncogene 2016, 35, 833–845. [Google Scholar] [CrossRef] [PubMed]
- Chaidos, A.; Caputo, V.; Gouvedenou, K.; Liu, B.; Marigo, I.; Chaudhry, M.S.; Rotolo, A.; Tough, D.F.; Smithers, N.N.; Bassil, A.K.; et al. Potent antimyeloma activity of the novel bromodomain inhibitors I-BET151 and I-BET762. Blood 2014, 123, 697–705. [Google Scholar] [CrossRef] [Green Version]
- Berenguer-Daize, C.; Astorgues-Xerri, L.; Odore, E.; Cayol, M.; Cvitkovic, E.; Noel, K.; Bekradda, M.; MacKenzie, S.; Rezai, K.; Lokiec, F.; et al. OTX015 (MK-8628), a novel BET inhibitor, displays in vitro and in vivo antitumor effects alone and in combination with conventional therapies in glioblastoma models. Int. J. Cancer 2016, 139, 2047–2055. [Google Scholar] [CrossRef]
- Picaud, S.; Da, C.D.; Thanasopoulou, A.; Filippakopoulos, P.; Fish, P.V.; Philpott, M.; Fedorov, O.; Brennan, P.; Bunnage, M.E.; Owen, D.R.; et al. PFI-1, a highly selective protein interaction inhibitor, targeting BET bromodomains. Cancer Res. 2013, 73, 3336–3346. [Google Scholar] [CrossRef] [Green Version]
- Wadhwa, E.; Nicolaides, T. Bromodomain inhibitor review: Bromodomain and extra-terminal family protein inhibitors as a potential new therapy in central nervous system tumors. Cureus 2016, 8, e620. [Google Scholar] [CrossRef] [Green Version]
- Cheng, X.; Huang, Z.; Long, D.; Jin, W. BET inhibitor bromosporine enhances 5-FU effect in colorectal cancer cells. Biochem. Biophys. Res. Commun. 2020, 521, 840–845. [Google Scholar] [CrossRef]
- Pérez-Salvia, M.; Esteller, M. Bromodomain inhibitors and cancer therapy: From structures to applications. Epigenetics 2017, 12, 323–339. [Google Scholar] [CrossRef]
- Albrecht, B.K.; Gehling, V.S.; Hewitt, M.C.; Vaswani, R.G.; Cote, A.; Leblanc, Y.; Nasveschuk, C.G.; Bellon, S.; Bergeron, L.; Campbell, R.; et al. Identification of a benzoisoxazoloazepine inhibitor (CPI-0610) of the bromodomain and extra-terminal (BET) family as a candidate for human clinical trials. J. Med. Chem. 2016, 59, 1330–1339. [Google Scholar] [CrossRef] [Green Version]
- Rvx 208. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3585949/ (accessed on 10 September 2019).
- Nicholls, S.J.; Puri, R.; Wolski, K.; Ballantyne, C.M.; Barter, P.J.; Brewer, H.B.; Kastelein, J.J.; Hu, B.; Uno, K.; Kataoka, Y.; et al. Effect of the BET protein inhibitor, RVX-208, on progression of coronary atherosclerosis: Results of the phase 2b, randomized, double-blind, multicenter, ASSURE trial. Am. J. Cardiovasc. Drugs 2016, 16, 55–65. [Google Scholar] [CrossRef] [PubMed]
- ClinicalTrials.gov. Available online: https://clinicaltrials.gov (accessed on 9 November 2016).
- Filippakopoulos, P.; Qi, J.; Picaud, S.; Shen, Y.; Smith, W.B.; Fedorov, O.; Morse, E.M.; Keates, T.; Hickman, T.T.; Felletar, I.; et al. Selective inhibition of BET bromodomains. Nature 2010, 468, 1067–1073. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Theodoulou, N.H.; Bamborough, P.; Bannister, A.J.; Becher, I.; Bit, R.A.; Che, K.H.; Chung, C.W.; Dittmann, A.; Drewes, G.; Drewry, DH.; et al. Discovery of I-BRD9, a selective cell active chemical probe for bromodomain containing protein 9 inhibition. J. Med. Chem. 2016, 59, 1425–1439. [Google Scholar] [CrossRef] [Green Version]
- Martin, L.J.; Koegl, M.; Bader, G.; Cockcroft, X.L.; Fedorov, O.; Fiegen, D.; Gerstberger, T.; Hofmann, M.H.; Hohmann, A.F.; Kessler, D.; et al. Structure-based design of an in vivo active selective BRD9 inhibitor. J. Med. Chem. 2016, 59, 4462–4475. [Google Scholar] [CrossRef]
- Hohmann, A.F.; Martin, L.J.; Minder, J.L.; Roe, J.S.; Shi, J.; Steurer, S.; Bader, G.; McConnell, D.; Pearson, M.; Gerstberger, T.; et al. Sensitivity and engineered resistance of myeloid leukemia cells to BRD9 inhibition. Nat. Chem. Biol. 2016, 12, 672–679. [Google Scholar] [CrossRef] [Green Version]
- Sachchidanand; Resnick-Silverman, L.; Yan, S.; Mutjaba, S.; Liu, W.J.; Zeng, L.; Manfredi, J.J.; Zhou, M.M. Target structure-based discovery of small molecules that block human p53 and CREB binding protein association. Chem. Biol. 2006, 13, 81–90. [Google Scholar] [CrossRef] [Green Version]
- Chekler, E.L.; Pellegrino, J.A.; Lanz, T.A.; Denny, R.A.; Flick, A.C.; Coe, J.; Langille, J.; Basak, A.; Liu, S.; Stock, IA.; et al. Transcriptional profiling of a selective CREB binding protein bromodomain inhibitor highlights therapeutic opportunities. Chem. Biol. 2015, 22, 1588–1596. [Google Scholar] [CrossRef]
- Picaud, S.; Fedorov, O.; Thanasopoulou, A.; Leonards, K.; Jones, K.; Meier, J.; Olzscha, H.; Monteiro, O.; Martin, S.; Philpott, M.; et al. Generation of a selective small molecule inhibitor of the CBP/p300 bromodomain for leukemia therapy. Cancer Res. 2015, 75, 5106–5119. [Google Scholar] [CrossRef] [Green Version]
- Pegg, N.; Brooks, N.; Worthington, J.; Young, B.; Prosser, A.; Lane, J.; Taddei, D.; Brown, R.; Harbottle, G.; Shannon, J.; et al. Characterisation of CCS1477: A novel small molecule inhibitor of p300/CBP for the treatment of castration resistant prostate cancer [abstract]. J. Clin. Oncol. 2017, 35 (Suppl. S15), 11590. [Google Scholar] [CrossRef]
- Sanchez, R.; Zhou, M.M. The PHD finger: A versatile epigenome reader. Trends Biochem. Sci. 2011, 36, 364–372. [Google Scholar] [CrossRef] [Green Version]
- Taverna, S.D.; Li, H.; Ruthenburg, A.J.; Allis, C.D.; Patel, D.J. How chromatin-binding modules interpret histone modifications: Lessons from professional pocket pickers. Nat. Struct. Mol. Biol. 2007, 14, 1025–1040. [Google Scholar] [CrossRef] [Green Version]
- Milosevich, N.; Hof, F. Chemical inhibitors of epigenetic methyllysine reader proteins. Biochemistry 2016, 55, 1570–1583. [Google Scholar] [CrossRef]
- Milosevich, N.; Gignac, M.C.; McFarlane, J.; Simhadri, C.; Horvath, S.; Daze, K.D.; Croft, C.S.; Dheri, A.; Quon, T.T.; Douglas, S.F.; et al. Selective inhibition of CBX6: A methyllysine reader protein in the Polycomb family. ACS Med. Chem. Lett. 2015, 7, 139–144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ali, M.; Daze, K.D.; Strongin, D.E.; Rothbart, S.B.; Rincon-Arano, H.; Allen, H.F.; Li, J.; Strahl, B.D.; Hof, F.; Kutateladze, T.G. Molecular insights into inhibition of the methylated histone-plant homeodomain complexes by calixarenes. J. Biol. Chem. 2015, 290, 22919–22930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wagner, E.K.; Nath, N.; Flemming, R.; Feltenberger, J.B.; Denu, J.M. Identification and characterization of small molecule inhibitors of a plant homeodomain finger. Biochemistry 2012, 51, 8293–8306. [Google Scholar] [CrossRef] [PubMed]
- Miller, T.C.; Rutherford, T.J.; Birchall, K.; Chugh, J.; Fiedler, M.; Bienz, M. Competitive binding of a benzimidazole to the histone-binding pocket of the Pygo PHD finger. ACS Chem. Biol. 2014, 9, 2864–2874. [Google Scholar] [CrossRef] [PubMed]
- Addou-Klouche, L.; Adélaïde, J.; Finetti, P.; Cervera, N.; Ferrari, A.; Bekhouche, I.; Sircoulomb, F.; Sotiriou, C.; Viens, P.; Moulessehoul, S.; et al. Loss, mutation and deregulation of L3MBTL4 in breast cancers. Mol. Cancer 2010, 9, 213. [Google Scholar] [CrossRef] [Green Version]
- Herold, J.M.; Wigle, T.J.; Norris, J.L.; Lam, R.; Korboukh, V.K.; Gao., C.; Ingerman, L.A.; Kireev, D.B.; Senisterra, G.; Vedadi, M.; et al. Small-molecule ligands of methyl-lysine binding proteins. J. Med. Chem. 2011, 54, 2504–2511. [Google Scholar] [CrossRef] [Green Version]
- Zhou, H.; Che, X.; Bao, G.; Wang, N.; Peng, L.; Barnash, K.D.; Frye, S.V.; James, L.I.; Bai, X. Design, synthesis, and protein methyltransferase activity of a unique set of constrained amine containing compounds. Bioorg. Med. Chem. Lett. 2016, 26, 4436–4440. [Google Scholar] [CrossRef] [Green Version]
- James, L.I.; Barsyte-Lovejoy, D.; Zhong, N.; Krichevsky, L.; Korboukh, V.K.; Herold, J.M.; MacNevin, C.J.; Norris, J.L.; Sagum, C.A.; Tempel, W.; et al. Discovery of a chemical probe for the L3MBTL3 methyllysine reader domain. Nat. Chem. Biol. 2013, 9, 184–191. [Google Scholar] [CrossRef]
- Perfetti, M.T.; Baughman, B.M.; Dickson, B.M.; Mu, Y.; Cui, G.; Mader, P.; Dong, A.; Norris, J.L.; Rothbart, S.B.; Strahl, B.D.; et al. Identification of a fragment-like small molecule ligand for the methyl-lysine binding protein, 53BP1. ACS Chem. Biol. 2015, 10, 1072–1081. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roatsch, M.; Hoffmann, I.; Abboud, M.I.; Hancock, R.L.; Tarhonskaya, H.; Hsu, K.F.; Wilkins, S.E.; Yeh, T.L.; Lippl, K.; Serrer, K.; et al. The clinically used iron chelator deferasirox is an inhibitor of epigenetic jumonjiC domain-containing histone demethylases. ACS Chem. Biol. 2019, 14, 1737–1750. [Google Scholar] [CrossRef] [PubMed]
- Baylin, S.B. DNA methylation and gene silencing in cancer. Nat. Clin. Pract. Oncol. 2005, 2 (Suppl. S1), S4–11. [Google Scholar] [CrossRef] [PubMed]
- Klauke, K.; Radulović, V.; Broekhuis, M.; Weersing, E.; Zwart, E.; Olthof, S.; Ritsema, M.; Bruggeman, S.; Wu, X.; Helin, K.; et al. Polycomb Cbx Family Members Mediate the Balance between Haematopoietic Stem Cell Self-Renewal and Differentiation. Nat. Cell Biol. 2013, 15, 353–362. [Google Scholar] [CrossRef]
- Morey, L.; Pascual, G.; Cozzuto, L.; Roma, G.; Wutz, A.; Benitah, S.A.; Di Croce, L. Nonoverlapping Functions of the Polycomb Group Cbx Family of Proteins in Embryonic Stem Cells. Cell Stem. Cell 2012, 10, 47–62. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.Z.; Chen, S.L.; Wang, C.H.; He, Y.F.; Yang, X.; Xie, D.; Yun, J.P. CBX8 exhibits oncogenic activity via AKT/b-Catenin activation in hepatocellular carcinoma. Cancer Res. 2017, 78, 51–63. [Google Scholar] [CrossRef] [Green Version]
- Chung, C.Y.; Sun, Z.; Mullokandov, G.; Bosch, A.; Qadeer, Z.A.; Cihan, E.; Rapp, Z.; Parsons, R.; Aguirre-Ghiso, J.A.; Farias, E.F.; et al. Cbx8 acts non-canonically with Wdr5 to promote mammary tumorigenesis. Cell Rep. 2016, 16, 472–486. [Google Scholar] [CrossRef] [Green Version]
- Tan, J.; Jones, M.; Koseki, H.; Nakayama, M.; Muntean, A.G.; Maillard, I.; Hess, J.L. CBX8, a polycomb group protein, is essential for MLL-AF9-induced leukemogenesis. Cancer Cell 2012, 20, 563–575. [Google Scholar] [CrossRef] [Green Version]
- Ren, C.; Morohashi, K.; Plotnikov, A.N.; Jakoncic, J.; Smith, S.G.; Li, J.; Zeng, L.; Rodriguez, Y.; Stojanoff, V.; Walsh, M.; et al. Small-molecule modulators of methyl-lysine binding for the CBX7 chromodomain. Chem. Biol. 2015, 22, 161–168. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Denton, K.E.; Hobbs, K.F.; Weaver, T.; McFarlane, J.M.B.; Connelly, K.E.; Gignac, M.C.; Milosevich, N.; Hof, F.; Paci, I.; et al. Optimization of ligands using focused DNA-encoded libraries to develop a selective, cell-permeable CBX8 chromodomain inhibitor. ACS Chem. Biol. 2020, 15, 112–131. [Google Scholar] [CrossRef]
- Stuckey, J.I.; Dickson, B.M.; Cheng, N.; Liu, Y.; Norris, J.L.; Cholensky, S.H.; Tempel, W.; Qin, S.; Huber, K.G.; Sagum, C.; et al. A cellular chemical probe targeting the chromodomains of Polycomb repressive complex 1. Nat. Chem. Biol. 2016, 12, 180–187. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Groups | Proteins | Name | BRDs | Alias | Functions and Recognize Histones |
|---|---|---|---|---|---|
| BRD I | BAZ1A | Bromodomain adjacent to zinc finger domain, 1A | 1 | ACF1, WALp1, WCRF180 | Chromatin remodeling factor [26] |
| BPTF | Fetal Alzheimer antigen | 1 | FALZ, FAC1 | Recognizes H4K16ac and H3K4me3 [27] | |
| CECR2 | Cat eye syndrome chromosome region, candidate 2 | 1 | KIAA1740 | Recognizes/binds to acetylated histones [18] | |
| GCN5L2 | General control of amino acid synthesis 5-like 2 | 1 | GCN5, KAT2A, STAF97, PCAF-B | HAT, interact with EP300/CREBBP and ADA2; Binds to H4K16ac [20,22] | |
| PCAF | P300/CBP-associated factor | 1 | CREBBP-associated factor, KAT2B | HAT, promotes transcriptional activation; Targets to H4K8ac, H3K14ac and H3K36ac [21,22] | |
| BRD II (BET) | BRD2 | Bromodomain-containing protein 2 | 2 | FSH, RING3 | Associated with acetylated chromatin during mitosis; Binds to the H4K5ac/K12ac and H3K14ac [28] |
| BRD3 | Bromodomain-containing protein 3 | 2 | ORFX, RING3L | Transcriptional regulator; Binds to the H4K5ac/K12ac and H3K14ac [28] | |
| BRD4 | Bromodomain-containing protein 4 | 2 | CAP, MCAP, HUNK1 | Interacts with acetylated H3K14 and H4K5/K8/K12/K16; Rerulates H3K27ac and H3K56ac [18,29] | |
| BRDT | Bromodomain-containing protein, testis specific | 2 | BRD6 | Chromatin remodeling factor; Recognizes H4K5ac/K8ac [30] | |
| BRD III | BAZ1B | Bromodomain adjacent to zinc finger domain, 1B | 1 | WSTF, WBSCR9 | Chromatin remodeling factor, transcriptional regulator; Recognizes H2BK12ac, H3K14ac and H4K16ac [31] |
| BRD8B | Bromodomain-containing protein 8 B | 2 | SMAP, SMAP2 | Transcriptional regulator [32] | |
| BRWD3 | Bromodomain-containing protein disrupted in leukemia | 2 | BRODL | Associated with translocations in patients with B-cell chronic lymphocytic leukemia [33] | |
| CREBBP | CREB Binding Protein | 1 | CBP, KAT3A | HAT; Binds to H4K20ac [22] | |
| EP300 | E1A-binding protein p300 | 1 | p300, KAT3B | HAT, acetylate H3K122/K27 and non-histone proteins [34,35] | |
| PHIP | Pleckstrin homology domain-interacting protein | 2 | DR11, WDR11, SRT1, HH14, BRWD2 | Binds to the insulin receptor substrate 1; Regulates growth and survival of pancreatic beta cells [36] | |
| WDR9 | WD repeat domain 9 | 2 | BRWD1 | Chromatin remodeling factor [37] | |
| BRD IV | ATAD2 | Two AAA domain containing protein | 1 | ANCCA | Transcriptional regulator; Binds to H3K14ac, H4K5ac/K12ac [38,39] |
| ATAD2B | KIAA1240 protein | 1 | KIAA1240 | Binds to acetylated chromatin [gene card] | |
| BRD1 | Bromodomain-containing protein 1 | 1 | BRPF2 | A subunit of the MOZ/MORF HAT complex, regulates H3K14ac [40] | |
| BRD7 | Bromodomain-containing protein 7 | 1 | BP75, NAG4, CELTIX1 | Transcriptional regulator; Binds to acetylated H3K9/K14, H4K8/K12/K16 [41] | |
| BRD9 | Bromodomain-containing protein 9 | 1 | LAVS3040, PRO9856 | Chromatin remodeling; Recognizes H4K5ac/K8ac [42] | |
| BRPF1 | Bromodomain- and PHD finger-containing protein 1A | 1 | BR140, Peregrin | Transcriptional activator; Recognizes acetylated H2AK5 and H3K14, H4K5/K8/K12 [43] | |
| BRPF3 | Bromodomain- and PHD finger-containing protein, 3 | 1 | KIAA1286 | Regulats replication origin activation and histone H3K14 acetylation [44] | |
| BRD V | BAZ2A | Bromodomain adjacent to zinc finger domain, 2A | 1 | TIP5, WALp3 | Transcriptional repressor; Interacts with H4K16ac [45] |
| BAZ2B | Bromodomain adjacent to zinc finger domain, 2B | 1 | WALp4 | Transcriptional regulator; Binds to H3K14ac [46] | |
| SP100 | Nuclear antigen Sp100 | 1 | Lysp100b | Binds heterochromatin and functions in immunity, and gene regulation [47] | |
| SP110 | Nuclear antigen Sp110 A, | 1 | IPR1, VODI, IFI41, IFI75 | Transcriptional activator [48] | |
| SP140 | SP140 nuclear body protein | 1 | LYSP100 | Associated with multiple sclerosis, Crohn’s disease, chronic lymphocytic [49,50] | |
| SP140L | SP140 nuclear body protein like | 1 | SP140L-1 protein | Chromatin binding [gene card] | |
| TRIM24 | Tripartite motif-containing 24 | 1 | TIF1a, PTC6, RNF82 | Transcriptional regulator; Recognizes H3K23ac [51] | |
| TRIM33 | Tripartite motif-containing 33 A | 1 | PTC7, RFG7, TIF1g | Control transcriptional elongation; Recognizes H3K18ac/K23ac [52] | |
| TRIM66 | Tripartite motif-containing 66 | 1 | TIF1d | Transcriptional repressor [53] | |
| BRD VI | MLL | Myeloid/lymphoid or mixed lineage leukemia | 1 | HRX, TRX1, CXXC7, ALL-1 | HMT, mediats H3K4me [23,25] |
| TRIM28 | Tripartite motif-containing 28 | 1 | KAP1, RNF96, TIF1b | Transcriptional regulator; Regulates H3K9ac and H3K14ac [gene card] | |
| BRD VII | BRWD3 | ||||
| PHIP | |||||
| TAF1 | TAF1 RNA polymerase II, TATA box-binding protein (TBP)-associated factor | 2 | TAFII250, P250, CCG1, TAF2A | Transcription inhibition; Binds to H4K5/K8/K12/K16ac [54] | |
| TAF1L | TAF1-like RNA polymerase II, TATA box-binding protein (TBP)-associated factor | 2 | TAF(II)210 | Functions as a TBP-associated factor [55] | |
| WDR9 | |||||
| ZMYND8 | Zinc Finger MYND-Type Containing 8 | 1 | PRKCBP1, RACK7 | Transcriptional regulator; Recognizes H4K5/K8/K12/K16/K20ac H3K9ac, and H3K14ac [56] | |
| ZMYND11 | remodeling factor containing 11 | 1 | BS69, BRAM1, MRD30 | Transcriptional repressor [57] | |
| BRD VIII | ASH1L | ash1 (absent, small, or homeotic)-like | 1 | ASH1, KMT2H | HMT, methylates H3K36me2 [24] |
| PBRM1 | Polybromo 1 | 6 | PB1, BAF180 | Chromatin remodeling factor; High-affinity with acetylated histone H3 at lysine 4, 9, 14 and 23 [58] | |
| SMARCA2 | SWI/SNF-related matrix associated actin-dependent regulator of chromatin a 2 | 1 | BRM, SNF2L2 | Chromatin remodeling factor, Splicing regulator; Interact with and moderate specificity for H3K14ac [59] | |
| SMARCA4 | SWI/SNF-related matrix associated actin-dependent regulator of chromatin a 4 | 1 | BRG1, SNF2L4, SNF2LB | Chromatin remodeling factor; Interact with and moderate specificity for H3K14ac [59] |
| Compounds | Conditions | Status | Clinical Trials Identifier |
|---|---|---|---|
| ABBV-075 | AML; Advanced Cancer; Breast Cancer; Multiple Myeloma; NHL; NSCLC; Prostate Cancer | Completed Phase 1 | NCT02391480 |
| BAY1238097 | Neoplasms | Terminated Phase 1 | NCT02369029 |
| BI 894999 | Neoplasms | Recruiting Phase 1 | NCT02516553 |
| BMS-986158 | Advanced Tumors | Recruiting Phase 1/2 | NCT02419417 |
| Childhood Solid Tumor; Lymphoma; Pediatric Brain Tumor | Recruiting Phase 1 | NCT03936465 | |
| CPI-0610 | AML; Myelofibrosis; MMN; MS | Recruiting Phase 1/2 | NCT02158858 |
| Lymphoma | Completed Phase 1 | NCT01949883 | |
| Multiple Myeloma | Completed Phase 1 | NCT02157636 | |
| Peripheral Nerve Tumors | Withdrawn Phase 2 | NCT02986919 | |
| FT-1101 | AML; Acute Myelogenous Leukemia; MS; NHL | Completed Phase 1 | NCT02543879 |
| GS-5829 | Advanced Estrogen Receptor; Positive HER2-Breast Cancer | Terminated Phase 1/2 | NCT02983604 |
| Lymphomas, Solid Tumors | Completed Phase 1 | NCT02392611 | |
| Metastatic CRPC | Active, not recruiting Phase 1/2 | NCT02607228 | |
| GSK2820151 | Solid Tumors | Active, not recruiting Phase 1 | NCT02630251 |
| GSK525762 (I-BET762) | Advanced and Retractory Solid Tumors; Lympnomas | Active, not recruiting Phase1 | NCT03925428 |
| Carcinoma Midline | Active, not recruiting Phase 1 | NCT01587703 | |
| Drug Interactions | Completed Phase 1 | NCT02706535 | |
| Neoplasms | Recruiting Phase 2 | NCT01943851 | |
| Neoplasms in combination with fulvestrant | Recruiting Phase 2 | NCT02964507 | |
| Solid Tumors | Recruiting Phase 1 | NCT03150056 | |
| Solid Tumours | Withdrawn Phase 2 | NCT03266159 | |
| Solid Tumours | Available | NCT03702036 | |
| INCB054329 | Hematologic Malignancy; Solid Tumors | Terminated Phase 1/2 | NCT02431260 |
| MK-8628 (OTX015) | AML | Active, not recruiting Phase 1 | NCT02698189 |
| AML | Withdrawn Phase 1/2 | NCT02303782 | |
| Acute Lymphoblastic Leukemia; AML; Diffuse Large B-cell Lymphoma; Multiple Myeloma | Completed Phase 1 | NCT01713582 | |
| Glioblastoma multiforme | Terminated Phase 2 | NCT02296476 | |
| CRPC; NUT Midline Carcinoma; NSCLC; Triple Negative Breast Cancer | Terminated Phase 1 | NCT02698176 | |
| CRPC; NUT Midline Carcinoma; NSCLC With Rearranged; ALK Gene/Fusion Protein or KRAS Mutation; Pancreatic Ductal Adenocarcinoma;Triple Negative Breast Cancer | Completed Phase 1 | NCT02259114 | |
| PLX51107 | AML; MS; NHL; Solid Tumors | Terminated Phase 1 | NCT02683395 |
| AML; MS; MMN; Myeloproliferative Neoplasm | Not yet recruiting Phase 1 | NCT04022785 | |
| RVX000222 | Fabry Disease | Not yet recruiting Phase 1/2 | NCT03228940 |
| Cardiovascular Diseases; Coronary Artery Disease; Type 2 Diabetes Mellitus | Active, not recruiting Phase3 | NCT02586155 | |
| TEN-010 (RO6870810) | Advances solid malignancies; Solid Tumors | Completed Phase 1 | NCT01987362 |
| AML; Myelodysplastic Syndromes | Completed Phase 1 | NCT02308761 | |
| Multiple Myeloma | Active, not recruiting Phase 1 | NCT03068351 | |
| ZEN003694 | Metastatic CRPC | Completed Phase 1 | NCT02705469 |
| Metastatic CRPC in combination with Enzalutamide | Active, not recruiting Phase 1/2 | NCT02711956 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, H.; Wei, T.; Cai, Y.; Jin, J. Small Molecules Targeting the Specific Domains of Histone-Mark Readers in Cancer Therapy. Molecules 2020, 25, 578. https://doi.org/10.3390/molecules25030578
Zhu H, Wei T, Cai Y, Jin J. Small Molecules Targeting the Specific Domains of Histone-Mark Readers in Cancer Therapy. Molecules. 2020; 25(3):578. https://doi.org/10.3390/molecules25030578
Chicago/Turabian StyleZhu, Huihui, Tao Wei, Yong Cai, and Jingji Jin. 2020. "Small Molecules Targeting the Specific Domains of Histone-Mark Readers in Cancer Therapy" Molecules 25, no. 3: 578. https://doi.org/10.3390/molecules25030578
APA StyleZhu, H., Wei, T., Cai, Y., & Jin, J. (2020). Small Molecules Targeting the Specific Domains of Histone-Mark Readers in Cancer Therapy. Molecules, 25(3), 578. https://doi.org/10.3390/molecules25030578