Relation between Crystal Structures of Precursors and Final Products: Example of Vitamin D Intermediates
Abstract
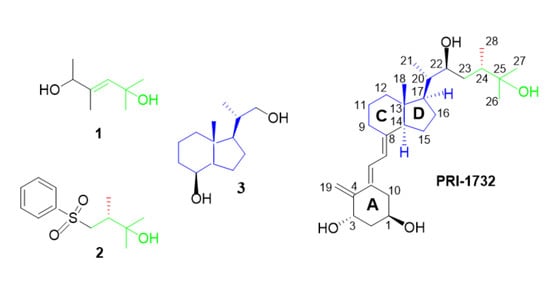
:1. Introduction
2. Results and Discussion
2.1. X-ray Diffraction Studies
2.2. Analogues of Vitamin D
2.3. Geometry Analysis
2.4. Intermolecular Interactions
2.4.1. Hydrogen Bonds Motifs
2.4.2. Hydrogen Bonds in the Intermediates
2.4.3. Hydrogen Bonds in the Intermediates and Vitamin D Analogues
2.4.4. Energy Frameworks
3. Materials and Methods
3.1. Synthesis and Crystallization
3.2. Single Crystal X-ray Studies
3.3. Dimer Calculations
3.4. CSD Search
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Okamura, W.H.; Norman, A.W.; Wing, R.M. Vitamin D: Concerning the Relationship between Molecular Topology and Biological Function. PNAS 1974, 71, 4194–4197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, A.J.; Slatopolsky, E. Vitamin D analogs: Therapeutic applications and mechanisms for selectivity. Mol. Asp. Med. 2008, 29, 433–452. [Google Scholar] [CrossRef] [PubMed]
- Bikle, D.D. Vitamin D Metabolism, Mechanism of Action, and Clinical Applications. Chem. Biol. 2014, 21, 319–329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boland, R. Role of Vitamin D in Skeletal Muscle Function. Endocr. Rev. 1986, 7, 434–448. [Google Scholar] [CrossRef] [PubMed]
- Marcinkowska, E.; Wallace, G.; Brown, G. The use of 1α, 25-dihydroxyvitamin D3 as an anticancer agent. Int. J. Mol. Sci. 2016, 17, 729. [Google Scholar] [CrossRef] [Green Version]
- Holick, M.F. Evolution and Function of Vitamin D. In Proceedings of the Vitamin D Analogs in Cancer Prevention and Therapy; Reichrath, J., Tilgen, W., Friedrich, M., Eds.; Springer: Berlin/Heidelberg, Germany, 2003; pp. 3–28. [Google Scholar]
- Holick, M.F. Vitamin D and Bone Health. J. Nutr. 1996, 126, 1159S–1164S. [Google Scholar] [CrossRef]
- DeLuca, H.F. Overview of general physiologic features and functions of vitamin D. Am. J. Clin. Nutr. 2004, 80, 1689S–1696S. [Google Scholar] [CrossRef] [Green Version]
- Christakos, S.; Hewison, M.; Gardner, D.G.; Wagner, C.L.; Sergeev, I.N.; Rutten, E.; Pittas, A.G.; Boland, R.; Ferrucci, L.; Bikle, D.D. Vitamin D: Beyond bone. Ann. N. Y. Acad. Sci. 2013, 1287, 45. [Google Scholar] [CrossRef]
- Sarkar, S.; Hewison, M.; Studzinski, G.P.; Li, Y.C.; Kalia, V. Role of vitamin D in cytotoxic T lymphocyte immunity to pathogens and cancer. Crit. Rev. Clin. Lab. Sci. 2016, 53, 132–145. [Google Scholar] [CrossRef]
- Leyssens, C.; Verlinden, L.; Verstuyf, A. The future of vitamin D analogs. Front. Physiol. 2014, 5, 122. [Google Scholar] [CrossRef] [Green Version]
- Scott, L.J.; Dunn, C.J.; Goa, K.L. Calcipotriol ointment. Am. J. Clin. Dermatol. 2001, 2, 95–120. [Google Scholar] [CrossRef] [PubMed]
- Peters, D.C.; Balfour, J.A. Tacalcitol. Drugs 1997, 54, 265–271. [Google Scholar] [CrossRef] [PubMed]
- Sprague, S.M.; Llach, F.; Amdahl, M.; Taccetta, C.; Batlle, D. Paricalcitol versus calcitriol in the treatment of secondary hyperparathyroidism. Kidney Int. 2003, 63, 1483–1490. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pietraszek, A.; Malińska, M.; Chodyński, M.; Krupa, M.; Krajewski, K.; Cmoch, P.; Woźniak, K.; Kutner, A. Synthesis and crystallographic study of 1,25-dihydroxyergocalciferol analogs. Steroids 2013, 78, 1003–1014. [Google Scholar] [CrossRef] [PubMed]
- Wanat, M.; Malinska, M.; Kutner, A.; Wozniak, K. Effect of vitamin D conformation on interactions and packing in the crystal lattice. Cryst. Growth Des. 2018, 18, 3385–3396. [Google Scholar] [CrossRef]
- Glebocka, A.; Sicinski, R.R.; Plum, L.A.; DeLuca, H.F. Ring-A-seco analogs of 1α,25-dihydroxy-19-norvitamin D3. J. Steroid Biochem. Mol. Biol. 2013, 136, 39–43. [Google Scholar] [CrossRef] [Green Version]
- Kolodziejski, W.; Woźniak, K.; Herold, J.; Dominiak, P.M.; Kutner, A. Crystal and molecular structure of 1α-hydroxylated analogs of vitamins D. J. Mol. Struct. 2005, 734, 149–155. [Google Scholar] [CrossRef]
- Groom, C.R.; Bruno, I.J.; Lightfoot, M.P.; Ward, S.C. The Cambridge structural database. Acta Crystallogr. Sect. B Struct. Sci. Cryst. Eng. Mater. 2016, 72, 171–179. [Google Scholar] [CrossRef]
- Allen, F.H. The Cambridge Structural Database: A quarter of a million crystal structures and rising. Acta Crystallogr. Sect. B Struct. Sci. 2002, 58, 380–388. [Google Scholar] [CrossRef]
- Toan, T.; Deluca, H.F.; Dahl, L.F. Solid-state conformations of vitamin D3. J. Org. Chem. 1976, 41, 3476–3478. [Google Scholar] [CrossRef]
- Hull, S.E.; Leban, I.; Main, P.; White, P.S.; Woolfson, M.M. The crystal and molecular structure of ergocalciferol (vitamin D2). Acta Crystallogr. Sect. B Struct. Crystallogr. Cryst. Chem. 1976, 32, 2374–2381. [Google Scholar] [CrossRef]
- Wang, J.-R.; Zhou, C.; Yu, X.; Mei, X. Stabilizing vitamin D 3 by conformationally selective co-crystallization. Chem. Commun. 2014, 50, 855–858. [Google Scholar] [CrossRef] [PubMed]
- Hatcher, M.A.; Peleg, S.; Dolan, P.; Kensler, T.W.; Sarjeant, A.; Posner, G.H. A-ring hydroxymethyl 19-nor analogs of the natural hormone 1α, 25-dihydroxyvitamin D3: Synthesis and preliminary biological evaluation. Bioorg. Med. Chem. 2005, 13, 3964–3976. [Google Scholar] [CrossRef] [PubMed]
- Posner, G.H.; Dai, H.; Afarinkia, K.; Murthy, N.N.; Guyton, K.Z.; Kensler, T.W. Asymmetric total synthesis, x-ray crystallography, and preliminary biological evaluation of 1-(1′-hydroxyethyl)-25-hydroxyvitamin D3 analogs of natural calcitriol. J. Org. Chem. 1993, 58, 7209–7215. [Google Scholar] [CrossRef]
- Hodgkin, D.C.; Rimmer, B.M.; Dunitz, J.D.; Trueblood, K.N. 947. The crystal structure of a calciferol derivative. J. Chem. Soc. 1963, 4945–4956. [Google Scholar] [CrossRef]
- Tan, E.S.; Tham, F.S.; Okamura, W.H. Vitamin D1. Chem. Commun. 2000, 23, 2345–2346. [Google Scholar] [CrossRef]
- Ryan, R.C.; Simon, G.L.; Calabrese, J.C.; Dahl, L.F.; DeLuca, H.F. Crystal structure of 25-hydroxy-vitamin D 3 monohydrate: A stereochemical analysis of vitamin D molecules. J. Chem. Soc. Perkin Trans. 1977, 2, 393–401. [Google Scholar]
- Bernstein, J.; Davis, R.E.; Shimoni, L.; Chang, N.-L. Patterns in Hydrogen Bonding: Functionality and Graph Set Analysis in Crystals. Angew. Chem. Int. Ed. Engl. 1995, 34, 1555–1573. [Google Scholar] [CrossRef]
- Brzeminski, P.; Fabisiak, A.; Sektas, K.; Berkowska, K.; Marcinkowska, E.; Sicinski, R.R. Synthesis of 19-norcalcitriol analogs with elongated side chain. J. Steroid Biochem. Mol. Biol. 2018, 177, 231–234. [Google Scholar] [CrossRef]
- Szybinski, M.; Brzeminski, P.; Fabisiak, A.; Berkowska, K.; Marcinkowska, E.; Sicinski, R.R. Seco-B-Ring Steroidal Dienynes with Aromatic D Ring: Design, Synthesis and Biological Evaluation. Int. J. Mol. Sci. 2017, 18, 2162. [Google Scholar] [CrossRef] [Green Version]
- Gándara, Z.; Suárez, P.L.; González, M.; Gómez, G.; Fall, Y. Vitamin D Heterocyclic Analogues; Part 2: Synthesis of the First Vitamin D Analogues with a Tetrazole Ring at the Side Chain. Synthesis 2011, 2011, 3887–3893. [Google Scholar]
- Wovkulich, P.M.; Barcelos, F.; Batcho, A.D.; Sereno, J.F.; Baggiolini, E.G.; Hennessy, B.M.; UskokoviĆ, M.R. Stereoselective total synthesis of 1α,25S,26-trihydroxycholecalciferol. Tetrahedron 1984, 40, 2283–2296. [Google Scholar] [CrossRef]
- Spek, A.L. CCDC 232012: Experimental Crystal Structure Determination. Camb. Crystallogr. Data Cent. (CCDC) 2004. [Google Scholar] [CrossRef]
- Martínez, A.; Gándara, Z.; González, M.; Gómez, G.; Fall, Y. Synthesis of new calcitriol analogues with an oxolane moiety in their side chains. Tetrahedron Lett. 2013, 54, 3514–3517. [Google Scholar] [CrossRef]
- Carballa, D.M.; Seoane, S.; Zacconi, F.; Pérez, X.; Rumbo, A.; Alvarez-Díaz, S.; Larriba, M.J.; Pérez-Fernández, R.; Muñoz, A.; Maestro, M.; et al. Synthesis and Biological Evaluation of 1α,25-Dihydroxyvitamin D3 Analogues with a Long Side Chain at C12 and Short C17 Side Chains. J. Med. Chem. 2012, 55, 8642–8656. [Google Scholar] [CrossRef] [PubMed]
- Nadkarni, S.; Chodyński, M.; Krajewski, K.; Cmoch, P.; Marcinkowska, E.; Brown, G.; Kutner, A. Convergent synthesis of double point modified analogs of 1α, 25-dihydroxyvitamin D2 for biological evaluation. J. Steroid Biochem. Mol. Biol. 2016, 164, 45–49. [Google Scholar] [CrossRef]
- Rigaku, O.D. CrysAlis PRO; Rigaku Oxford Diffraction: Yarnton, UK, 2015. [Google Scholar]
- Dolomanov, O.V.; Bourhis, L.J.; Gildea, R.J.; Howard, J.A.; Puschmann, H. OLEX2: A complete structure solution, refinement and analysis program. J. Appl. Crystallogr. 2009, 42, 339–341. [Google Scholar] [CrossRef]
- Sheldrick, G.M. A short history of SHELX. Acta Crystallogr. Sect. A Found. Crystallogr. 2008, 64, 112–122. [Google Scholar] [CrossRef] [Green Version]
- Sheldrick, G.M. Crystal structure refinement with SHELXL. Acta Cryst. C 2015, 71, 3–8. [Google Scholar] [CrossRef]
- Prince, E.; Spiegelman, C.H. International Tables for Crystallography; Kluwer Academic Publishers: Dordrecht, The Nederland, 1992; Volume C, pp. 622–624. [Google Scholar]
- Turner, M.J.; Thomas, S.P.; Shi, M.W.; Jayatilaka, D.; Spackman, M.A. Energy frameworks: Insights into interaction anisotropy and the mechanical properties of molecular crystals. Chem. Commun. 2015, 51, 3735–3738. [Google Scholar] [CrossRef]
- Turner, M.J.; McKinnon, J.J.; Wolff, S.K.; Grimwood, D.J.; Spackman, P.R.; Jayatilaka, D.; Spackman, M.A. Crystal Explorer17; University of Western Australia: Crawley WA, Australia, 2017. [Google Scholar]
- Gavezzotti, A. Non-conventional bonding between organic molecules. The ‘halogen bond’ in crystalline systems. Mol. Phys. 2008, 106, 1473–1485. [Google Scholar] [CrossRef] [Green Version]
- Jarzembska, K.N.; Dominiak, P.M.; Messerschmidt, M.; Volkov, A. LSDB-Program for Automatic Assignment of Local Coordinate Systems and UBDB Transfer. Available online: http://crystal.chem.uw.edu.pl/software/ubdb.html, (accessed on 17 October 2017).
- Jarzembska, K.N.; Dominiak, P.M.; Li, X.; Volkov, A. PseudoAtom Multipolar DataBank. Available online: http://crystal.chem.uw.edu.pl/software/ubdb.html (accessed on 17 October 2017).
- Spencer, T.A.; Li, D.; Russel, J.S.; Tomkinson, N.C.O.; Willson, T.M. Further Studies on the Synthesis of 24(S),25-Epoxycholesterol. A New, Efficient Preparation of Desmosterol. J. Org. Chem. 2000, 65, 1919–1923. [Google Scholar] [CrossRef] [PubMed]
- Day, R.O.; Kingsbury, C.A.; Day, V.W. Conformational preferences of tetrasubstituted ethanes. Steric perturbations in sulfones vs. sulfoxides. J. Org. Chem. 1981, 46, 1004–1009. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are not available from the authors. |








| PRI-1730 | PRI-1731 | PRI-1732 | 1S | 1R | 2 | |
|---|---|---|---|---|---|---|
| C25-C26 | 1.519 (6) | 1.523 (4) | 1.524 (3) | 1.523 (3) | 1.523 (3) | 1.506 (6) |
| C25-C27 | 1.522 (6) | 1.523 (4) | 1.524 (3) | 1.528 (3) | 1.529 (3) | 1.531 (5) |
| C25-O25 | 1.447 (5) | 1.436 (3) | 1.452 (3) | 1.447 (2) | 1.445 (2) | 1.441 (5) |
| C25-C24 | 1.556 (6) | 1.549 (4) | 1.552 (3) | 1.515 (3) | 1.520 (3) | 1.536 (5) |
| C24-C28 | 1.538 (6) | 1.530 (4) | 1.532 (3) | n/a | n/a | 1.530 (5) |
| C24-C23 | 1.538 (5) | 1.500 (3) | 1.537 (2) | n/a | n/a | 1.542 (5) |
| PRI-1730 | PRI-1731 | PRI-1732 | 3 | |
|---|---|---|---|---|
| C13-C17 | 1.558 (5) | 1.558 (2) | 1.561 (2) | 1.563 (3) |
| C13-C18 | 1.537 (5) | 1.529 (3) | 1.533 (2) | 1.524 (4) |
| C13-C14 | 1.563 (5) | 1.554 (3) | 1.546 (2) | 1.549 (4) |
| C13-C12 | 1.541 (5) | 1.527 (3) | 1.536 (2) | 1.526 (4) |
| C22-C20 | 1.546 (5) | 1.503 (3) | 1.548 (2) | 1.527 (4) |
| C17-C20 | 1.552 (5) | 1.540 (3) | 1.540 (2) | 1.538 (4) |
| C17-C16 | 1.552 (5) | 1.550 (3) | 1.563 (2) | 1.554 (4) |
| C20-C21 | 1.532 (6) | 1.526 (3) | 1.518 (3) | 1.516 (4) |
| C14-C8 | 1.509 (5) | 1.507 (3) | 1.506 (2) | 1.534 (4) |
| C14-C15 | 1.522 (5) | 1.524 (3) | 1.516 (2) | 1.516 (6) |
| C8-C9 | 1.514 (5) | 1.510 (3) | 1.507 (3) | 1.512 (7) |
| C11-C12 | 1.526 (5) | 1.537 (3) | 1.543 (2) | 1.543 (4) |
| C11-C9 | 1.536 (6) | 1.528 (3) | 1.533 (3) | 1.525 (7) |
| C16-C15 | 1.561 (5) | 1.545 (3) | 1.545 (2) | 1.556 (4) |
| Molecule | Hydrogen Bonds | Length of Hydrogen Bonds [Å] | Coulomb Energy [kJ/mol] | Dispersion Energy [kJ/mol] | Total Energy [kJ/mol] | |
|---|---|---|---|---|---|---|
| 1 | O2-H2…O5A | HB10a | 1.968 (1) | −47.6 | −19.7 | −35.8 |
| O5A-H5AA…O2A | HB11a | 1.916 (2) | −46.4 | −19.5 | −43.5 | |
| O2A-H2AA…O5 | HB10b | 1.937 (2) | −49.2 | −27.1 | −43.7 | |
| O5-H5…O2 | HB11b | 1.930 (2) | −46.4 | −19.5 | −43.5 | |
| 2 | O3-H3…O1 | HB12 | 2.011 (3) | −34.1 | −14.3 | −30.3 |
| Molecule | Hydrogen Bonds | Geometrical Similarity | Energetic Similarity | |||
|---|---|---|---|---|---|---|
| Coulomb Energy | Dispersion Energy | Total Energy | ||||
| 1 | O2-H2…O5A | HB10a | HB2 | HB2/HB3 | HB6 | HB3/HB6 |
| O5A-H5AA…O2A | HB11a | HB2, HB3 | HB3 | HB6 | - | |
| O2A-H2AA…O5 | HB10b | HB3, HB8 | HB2 | - | - | |
| O5-H5…O2 | HB11b | HB6 | HB3 | HB6 | - | |
| 2 | O3-H3…O1 | HB12 | HB7, HB1 | HB1 | HB3 | HB2 |
| Crystal Data | |||
|---|---|---|---|
| 1 | 2 | 3 | |
| Chemical formula | C8H16O2 | C12H18O3S | C9.75H17.75O1.5 |
| Mr | 144.21 | 242.32 | 158.99 |
| Crystal system, space group | Monoclinic, P21/c | Orthorhombic, P212121 | Monoclinic, C2 |
| Temperature (K) | 100 | 100 | 100 |
| a, b, c (Å) | 20.4858 (5), 6.02000 (12), 15.3758 (4) | 5.63736 (9), 7.88326 (9), 28.8351 (4) | 18.59 (3), 6.891 (7), 15.65 (2) |
| β (°) | 111.124 (3) | 107.53 (16) | |
| V (Å3) | 1768.80 (7) | 1281.45 (3) | 1912 (5) |
| Z | 8 | 4 | 8 |
| Radiation type | Cu Kα | ||
| µ (mm−1) | 0.61 | 2.18 | 0.56 |
| Crystal size (mm) | 0.39 × 0.1 × 0.09 | 0.27 × 0.22 × 0.15 | 0.23 × 0.07 × 0.06 |
| Data Collection | |||
| Diffractometer | SuperNova, Dual, Cu at zero, Atlas | ||
| Absorption correction | Multi-scan CrysAlis PRO 1.171.38.41 (Rigaku Oxford Diffraction, 2015) Empirical absorption correction using spherical harmonics, implemented in SCALE3 ABSPACK scaling algorithm. | ||
| Tmin, Tmax | 0.784, 1.000 | 0.775, 1.000 | 0.808, 1.000 |
| No. of measured, independent and observed [I>2σ(I)] reflections | 13,249, 3712, 3654 | 12,735, 2669, 2636 | 19,999, 3648, 3425 |
| Rint | 0.028 | 0.027 | 0.033 |
| (sin θ/λ)max (Å−1) | 0.632 | 0.631 | 0.617 |
| Refinement | |||
| R[F2 > 2σ(F2)], wR(F2), S | 0.066, 0.162, 1.15 | 0.045, 0.107, 1.10 | 0.0427, 0.109, 1.07 |
| No. of reflections | 3712 | 2669 | 3684 |
| No. of parameters | 193 | 149 | 343 |
| No. of restraints | 0 | 0 | 23 |
| H-atom treatment | H-atom parameters constrained | H-atoms treated by a mixture of independent and constrained refinement | |
| Δρmax, Δρmin (e Å−3) | 0.32, −0.19 | 0.43, −0.53 | 0.18, −0.23 |
| Absolute structure | n/a | Flack x determined using 1024 quotients [(I+)−(I−)]/[(I+)+(I−)] (Parsons, Flack and Wagner, Acta Cryst. B69 (2013) 249–259). | Absolute structure: Flack x determined using 1428 quotients [(I+)−(I−)]/[(I+)+(I−)] (Parsons, Flack and Wagner, Acta Cryst. B69 (2013) 249–259). |
| Absolute structure parameter | n/a | 0.011 (8) | −0.06 (8) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wanat, M.; Malinska, M.; Kutner, A.; Woźniak, K. Relation between Crystal Structures of Precursors and Final Products: Example of Vitamin D Intermediates. Molecules 2020, 25, 1802. https://doi.org/10.3390/molecules25081802
Wanat M, Malinska M, Kutner A, Woźniak K. Relation between Crystal Structures of Precursors and Final Products: Example of Vitamin D Intermediates. Molecules. 2020; 25(8):1802. https://doi.org/10.3390/molecules25081802
Chicago/Turabian StyleWanat, Monika, Maura Malinska, Andrzej Kutner, and Krzysztof Woźniak. 2020. "Relation between Crystal Structures of Precursors and Final Products: Example of Vitamin D Intermediates" Molecules 25, no. 8: 1802. https://doi.org/10.3390/molecules25081802
APA StyleWanat, M., Malinska, M., Kutner, A., & Woźniak, K. (2020). Relation between Crystal Structures of Precursors and Final Products: Example of Vitamin D Intermediates. Molecules, 25(8), 1802. https://doi.org/10.3390/molecules25081802