5-(1H-Indol-3-ylmethylene)-4-oxo-2-thioxothiazolidin-3-yl)alkancarboxylic Acids as Antimicrobial Agents: Synthesis, Biological Evaluation, and Molecular Docking Studies
Abstract
:1. Introduction

2. Results and Discussion
2.1. Chemistry
2.2. Biological Evaluation
2.2.1. Antibacterial Activity
2.2.2. Antifungal Activity
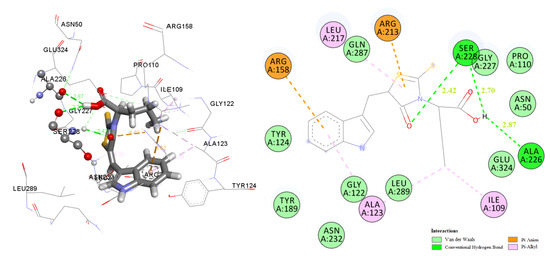
2.3. Docking Studies
2.3.1. Docking to Antibacterial Targets
2.3.2. Docking to Lanosterol 14α-Demethylase of C. albicans
3. Experimental Part
3.1. General Procedure for the Synthesis of 4-Oxo-2-thioxothiazolidin-3-ylalkanecarboxylic Acids 2a–d and 3a–g
3.2. General Procedure for the Synthesis of 5-(1-R1,5-R2,6-R3-1H-Indol-3-ylmethylene)-4-oxo-2- thioxothiazolidin-3-yl] Alkane Carboxylic Acids 4a–i and 5a–k
3.3. Biological Evaluation
3.3.1. Antibacterial Activity
3.3.2. Inhibition of Biofilm Formation
3.3.3. Antifungal Activity
3.4. Docking Studies
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Nii-Trebi, N.I. Emerging and neglected infectious diseases: Insights, advances, and challenges. Biomed. Res. Int. 2017, 2017, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livorsi, D.J.; Stenehjem, E.; Stephens, D.S. Sepsis -Pro-Inflammatory and Anti-Inflammatory Responses; Herwald, H., Egesten, A., Eds.; Karger Publishers: Basel, Switzerland, 2011; Volume 17, pp. 31–48. [Google Scholar]
- Shuvankar Mukherjee, S. Emerging infectious diseases: Epidemiological perspective. Indian J. Dermatol. 2017, 62, 459–467. [Google Scholar] [PubMed]
- Holmes, A.; Moore, L.; Sundsfjord, A.; Steinbakk, M.; Regmi, S.; Karkey, A.; Guerin, P.; Piddock, L. Understanding the mechanisms and drivers of antimicrobial resistance. Lancet 2016, 387, 176–187. [Google Scholar] [CrossRef]
- Winter, S.E.; Lopez, C.A.; Bäumler, A.J. The dynamics of gut-associated microbial communities during inflammation. EMBO Rep. 2013, 14, 319–327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Payne, D.J.; Gwynn, M.N.; Holmes, D.J.; Pompliano, D.L. Drugs for bad bugs: Confronting the challenges of antibacterial. Nat. Rev. Drug Discov. 2007, 6, 29–40. [Google Scholar] [CrossRef]
- He, X.Y.; Lu, L.; Qiu, J.; Zou, P.; Yu, F.; Jiang, X.K.; Li, L.; Jiang, S.; Liu, S.; Xie, L. Small molecule fusion inhibitors: Design, synthesis and biological evaluation of (Z)-3-(5-(3-benzyl-4-oxo-2-thioxothiazolidinylidene)methyl)-N-(3-carboxy-4-hydroxy)phe-nyl-2,5-dimethylpyrroles and related derivatives targeting HIV-1gp41. Bioorg. Med. Chem. 2013, 21, 7539–7548. [Google Scholar] [CrossRef] [PubMed]
- Yingchoncharoen, P.; Kalinowski, D.S.; Richardson, D.R. Lipid-baseddrug delivery systems in cancer therapy: What is available andwhat is yet to come. Pharmacol. Rev. 2016, 68, 701–787. [Google Scholar] [CrossRef] [Green Version]
- Nitsche, C.; Schreier, V.N.; Behnam, M.A.M. Thiazolidinone–peptide hybrids as dengue virus protease inhibi-tors with antiviral activity in cell culture. J. Med. Chem. 2013, 56, 8389–8403. [Google Scholar] [CrossRef]
- Bari, S.B.; Firake, S.D. Exploring anti-inflammatory Potential of thiazolidinone derivatives of benzenesulfonamide via synthesis, molecular docking and biological evaluation. Anti-Inflamm. Anti-Allergy Agents Med. Chem. 2016, 15, 44–53. [Google Scholar] [CrossRef]
- Khaled, R.A.; Mohamed, A.A.; Heba, A.H.; Shahinda, S.R. Design, synthesis and biological screening of new 4-thiazolidinone derivatives with promising COX-2 selectivity, anti-inflammatory activity and gastric safety profile. Bioorg. Chem. 2016, 64, 1–12. [Google Scholar]
- Yasmin, S.; Capone, F.; Laghezza, A.; Dal Piaz, F.; Loiodice, F.; Vijayan, V.; Devadasan, V.; Mondal, S.; Atlı, O.; Baysal, M.; et al. Novel benzylidene thiazolidinedione derivatives as partial PPARγ agonists and their antidiabetic effects on type 2 diabetes. Sci. Rep. 2017, 7, 14453. [Google Scholar] [CrossRef] [PubMed]
- Djukic, M.; Fesatidou, M.; Xenikakis, I.; Geronikaki, A.; Angelova, V.; Savic, V.; Pasic, M.; Krilovic, B.; Djukic, D.; Gobeljic, B.; et al. In vitro antioxidant activity of thiazolidinone derivatives of 1,3-thiazole and 1,3,4-thiadiazole. Chem. Biol. Interact. 2018, 286, 119–131. [Google Scholar] [CrossRef] [PubMed]
- Shafii, N.; Khoobi, M.; Amini, M. Synthesis and biologicalevaluation of 5-benzylidenerhodanine-3-acetic acid derivatives asAChE and 15-LOX inhibitors. J. Enzym. Inhib. Med. Chem. 2015, 30, 389–395. [Google Scholar] [CrossRef] [Green Version]
- Kratky, M.; Stepankova, S.; Vorcakova, K.; Vinšová, J. Synthesis and invitro evaluation of novel rhodanine derivatives as potential cho-linesterase inhibitors. Bioorg. Chem. 2016, 68, 23–29. [Google Scholar] [CrossRef] [PubMed]
- El-Sayed, S.; Metwally, K.; El-Shanawani, A.A.; Abdel-Aziz, L.M.; El-Rashedy, A.A.; Soliman, M.E.S.; Quattrini, L.; Coviello, V.; la Motta, C. Quinazolinone-based rhodanine-3-acetic acids as potent aldose reductase inhibitors: Synthesis, functional evaluation and molecular modeling study. Bioorg. Med. Chem. Lett. 2017, 27, 4760–4764. [Google Scholar] [CrossRef] [PubMed]
- Sim, M.M.; Ng, S.B.; Buss, A.D. Benzylidene rhodanines as novel inhibitors of UDP-N-acetylmuramate/l-alanine ligase. Bioorg. Med. Chem. Lett. 2002, 12, 697–699. [Google Scholar] [CrossRef]
- Miao, J.; Zheng, C.-J.; Sun, L.-P.; Song, M.-X.; Xu, L.-L.; Piao, H.-R. Synthesis and potential anti-bacterial activity of new rhodanine-3-acetic acid derivatives. Med. Chem. Res. 2013, 22, 4125–4132. [Google Scholar] [CrossRef]
- Gupta, A.; Singh, R.; Sonar, P.K.; Saraf, S.K. Novel 4-thiazolidinone deriva-tives as anti-infective agents: Synthesis, characterization, andantimicrobial evaluation. Biochem. Res. Int. 2016, 2016, 1–8. [Google Scholar]
- Tejchman, W.; Korona-Glowniak, I.; Malm, A.; Zylewski, M.; Suder, P. Antibacterialproperties of 5-substituted derivatives of rhodanine-3-carbox-yalkyl acids. Med. Chem. Res. 2017, 26, 1316–1324. [Google Scholar] [CrossRef] [Green Version]
- Song, M.X.; Zheng, C.J.; Deng, X.Q.; Wei, Z.-Y.; Piao, H.-R. The synthesis and anti-bacterial activities of N-carboxymethyl rhodanines. Med. Chem. 2014, 4, 441–448. [Google Scholar]
- Krátký, M.; Vinšová, J.; Stolaříková, J. Antimicrobial activity of rhodanine-3-acetic acid derivatives. Bioorg. Med. Chem. 2017, 25, 1839–1845. [Google Scholar] [CrossRef] [PubMed]
- Zheng, C.J.; Song, M.X.; Sun, L.P.; Wu, Y.; Hong, L.; Piao, H.R. Synthesis and biological evalution of 5-aryloxypyrazole derivatives bearing a rhodanine-3-aromatic acid as potential antimicrobial agents. Bioorg. Med. Chem. Lett. 2012, 22, 7024–7028. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Zhai, X.; Zhong, Z.; Li, G.; Pu, Y.; Gong, P. Design, synthesis and evaluation of novel rhodanine-containing sorafenib analogs as potential antitumor agents. Arch. Pharm. (Weinheim) 2011, 344, 349–357. [Google Scholar] [CrossRef]
- Lafayette, E.A.; de Almeida, S.M.V.; Santos, R.V.C.; de Oliveira, J.F.; da Cruz Amorim, C.A.; da Silva, R.M.F.; da Rocha Pitta, M.G.; da Rocha Pitta, I.; de Moura, R.O.; de Carvalho Junior, L.B.; et al. Synthesis of novel indole derivatives as promising DNA-binding agents and evaluation of antitumor and antitopoisomerase I activities. Eur. J. Med. Chem. 2017, 136, 511–522. [Google Scholar] [CrossRef] [PubMed]
- Villain-Guillot, P.; Gualtieri, M.; Bastide, L.; Roquet, F.; Martinez, J.; Amblard, M.; Pugniere, M.; Leonetti, J.P. Structure-activity relationships of phenyl-furanyl-rhodanines as inhibitors of RNA polymerase with antibacterial activity on biofilms. J. Med. Chem. 2007, 50, 4195–4204. [Google Scholar] [CrossRef] [PubMed]
- Song, M.X.; Li, S.H.; Peng, J.Y.; Guo, T.T.; Xu, W.H.; Xiong, S.F.; Deng, X.Q. Synthesis and bioactivity evaluation of N-arylsulfonylindole analogs bearing a rhodanine moiety as antibacterial agents. Molecules 2017, 22, 970. [Google Scholar] [CrossRef] [Green Version]
- Johnson, S.L.; Chen, L.; Harbach, R.; Sabet, M.; Savinov, A.; Cotton, N.J.; Strongin, A.; Guiney, D.; Pellecchia, M. Rhodanine derivatives as selective protease inhibitors against bacterial toxins. Chem. Biol. Drug Des. 2008, 71, 131–139. [Google Scholar] [CrossRef]
- Bataille, C.J.; Brennan, M.B.; Byrne, S.; Davies, S.G.; Durbin, M.; Fedorov, O.; Huber, K.; Jones, A.; Knapp, S.; Liu, G.; et al. Thiazolidine derivatives as potent and selective inhibitors of the PIM kinase family. Bioorg. Med. Chem. 2017, 25, 2657–2665. [Google Scholar] [CrossRef]
- Pinson, J.A.; Schmidt-Kittler, O.; Zhu, J.; Jennings, I.G.; Kinzler, K.W.; Vogelstein, B.; Chalmers, D.K.; Thompson, P.E. Thiazolidinedione-based PI3Kα inhibitors: An Analysis of biochemical and virtual screening methods. Chem. Med. Chem. 2011, 6, 514–522. [Google Scholar] [CrossRef]
- Song, H.; Lee, Y.S.; Roh, E.J.; Seo, J.H.; Oh, K.S.; Lee, B.H.; Han, H.; Shin, K.J. Discovery of potent and selective rhodanine type IKKβ inhibitors by hit-to-lead strategy. Bioorg. Med. Chem. Lett. 2012, 22, 5668–5674. [Google Scholar] [CrossRef]
- Sukanta, K.; Edward, B.R. Microwave-assisted synthesis of novel bis(2-thioxothiazolidin-4-one) derivatives as potential GSK-3 inhibitors. Tetrahedron Lett. 2012, 53, 3998–4003. [Google Scholar]
- Parrino, B.; Diana, P.; Cirrincione, G.; Casciofero, S. Bacterial biofilm inhibition in the development of effective anti-virulence strategy. Open Med. Chem. J. 2018, 12, 84–87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kostić, M.; Smiljković, M.; Petrović, J.; Glamočilija, J.; Barros, L.; Ferreira, I.C.F.R.; Ćirić, A.; Soković, M. Chemical, nutritive composition and a wide range of bioactive properties of honey mushroom Armillaria mellea (Vahl: Fr.) Kummer. Food Funct. 2017, 8, 3239–3249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clinical and Laboratory Standards Institute. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically. Approved Standard, 8th ed.; CLSI publication M07-A8; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2009. [Google Scholar]
- Fesatidou, M.; Zagaliotis, P.; Camoutsis, C.; Petrou, A.; Eleftheriou, P.; Tratrat, C.; Haroun, M.; Geronikaki, A.; Ciric, A.; Sokovic, M. 5-Adamantan thiadiazole- based thiazolidinones as antimicrobial agents. Design, synthesis, molecular docking and evaluation. Bioorg. Med. Chem. 2018, 26, 4664–4676. [Google Scholar] [CrossRef] [PubMed]
- Kartsev, V.; Geronikak, A.; Petrou, A.; Lichitsky, B.; Kostic, M.; Smiljkovic, M.; Soković, M.; Sirakanyan, S. Griseofulvin derivatives: Synthesis, molecular docking and biological evaluation. Curr. Top. Med. Chem. 2019, 19, 1145–1161. [Google Scholar] [CrossRef]
- Espinel-Ingroff, A. Comparison of the E-test with the NCCLS M38-P method for antifungal susceptibility testing of common and emerging pathogenic filamentous fungi. J. Clin. Microb. 2001, 39, 1360–1367. [Google Scholar] [CrossRef] [Green Version]
- Hanel, H.; Raether, W. A More Sophisticated Method of Determining the Fungicidal Effect of Water-Insoluble Preparations with a Cell Harvester, Using Miconazole as an Example. MYCOSES 1988, 31, 148–154. [Google Scholar] [CrossRef]
- Drenkard, E.; Ausubel, F.M. Pseudomonas biofilm formationand antibiotic resistance are linked to phenotypic variation. Nature 2002, 416, 740–743. [Google Scholar] [CrossRef]
Sample Availability: Some samples of the compounds are available from the authors. |




| R.br | B.c | M.f | S.a | L.m | En.cl | P.a | S.t | E. coli | |
|---|---|---|---|---|---|---|---|---|---|
| 4a | MIC | 4.72 | 28.30 | 6.92 | 9.43 | 4.72 | 4.72 | 4.72 | 28.30 |
| MBC | 9.43 | 37.72 | 9.43 | 18.86 | 9.43 | 9.43 | 9.43 | 37.72 | |
| 4b | MIC | 4.51 | 13.54 | 2.26 | 9.03 | 2.26 | 2.26 | 18.05 | 7.47 |
| MBC | 9.03 | 18.05 | 4.51 | 18.05 | 4.51 | 4.51 | 36.10 | 18.05 | |
| 4c | MIC | 8.61 | 12.91 | 8.61 | 8.61 | 4.31 | 4.31 | 4.31 | 12.91 |
| MBC | 17.22 | 17.22 | 17.22 | 17.22 | 8.61 | 8.61 | 8.61 | 17.22 | |
| 4d | MIC | 8.66‘ | 4.33 | 8.66 | 8.66‘ | 4.33 | 4.33 | 8.66‘ | 8.66‘ |
| MBC | 17.32 | 8.66‘ | 17.32 | 17.32 | 8.66 | 8.66‘ | 17.32 | 17.32 | |
| 4e | MIC | 8.28 | 4.14 | 4.14 | 8.28 | 8.28 | 4.14 | 8.28 | 12.42 |
| MBC | 16.57 | 8.28 | 8.28 | 16.57 | 16.57 | 8.28 | 16.57 | 16.57 | |
| 4f | MIC | 4.16 | 8.32 | 4.16 | 16.64 | 4.16 | 4.16 | 16.64 | 8.32 |
| MBC | 8.32 | 16.64 | 8.32 | 33.2 | 8.32 | 8.32 | 33.29 | 16.64 | |
| 4g | MIC | 7.97 | 3.98 | 3.98 | 11.95 | 3.98 | 3.98 | 14.07 | 1.59 |
| MBC | 15.94 | 7.97 | 7.97 | 15.94 | 7.97 | 7.97 | 15.94 | 1.99 | |
| 4h | MIC | 3.98 | 1.99 | 1.99 | 3.98 | 3.98 | 3.98 | 3.98 | 3.98 |
| MBC | 7.97 | 3.98 | 3.98 | 7.97 | 7.97 | 7.97 | 7.97 | 7.97 | |
| 4i | MIC | 4.00 | 5.87 | 2.94 | 16.02 | 5.87 | 5.87 | 16.02 | 5.87 |
| MBC | 8.01 | 8.01 | 4.00 | 32.04 | 8.01 | 8.01 | 32.04 | 8.01 | |
| 5a | MIC | 6.63 | 18.07 | 6.63 | 9.04 | 3.31 | 4.52 | 9.04 | 18.07 |
| MBC | 9.04 | 36.14 | 9.04 | 18.07 | 4.52 | 9.04 | 18.07 | 36.14 | |
| 5b | MIC | 1.11 | 12.50 | 0.56 | 2.22 | 1.67 | 2.22 | 2.22 | 4.17 |
| MBC | 2.08 | 16.67 | 2.08 | 4.17 | 2.08 | 8.33 | 4.17 | 8.33 | |
| 5c | MIC | 7.64 | 3.82 | 7.64 | 7.64 | 3.82 | 7.64 | 7.64 | 3.82 |
| MBC | 15.29 | 7.64 | 15.29 | 15.29 | 7.64 | 15.29 | 15.29 | 7.64 | |
| 5d | MIC | 3.69 | 3.69 | 3.69 | 7.38 | 3.69 | 3.69 | 7.38 | 3.69 |
| MBC | 7.38 | 7.38 | 7.38 | 14.76 | 7.38 | 7.38 | 14.76 | 7.38 | |
| 5e | MIC | 7.10 | 5.21 | 7.10 | 7.10 | 3.55 | 3.55 | 3.55 | 7.10 |
| MBC | 14.20 | 7.10 | 14.20 | 14.20 | 7.10 | 7.10 | 7.10 | 14.20 | |
| 5f | MIC | 11.02 | 11.02 | 5.39 | 7.34 | 5.39 | 5.39 | 14.69 | 11.02 |
| MBC | 14.69 | 14.69 | 7.34 | 14.69 | 7.34 | 7.34 | 29.38 | 14.69 | |
| 5g | MIC | 1.96 | 11.03 | 1.96 | 1.96 | 3.68 | 1.96 | 1.96 | 11.03 |
| MBC | 3.68 | 14.72 | 3.68 | 3.68 | 7.36 | 3.68 | 3.68 | 14.72 | |
| 5h | MIC | 7.10 | 7.10 | 3.55 | 7.10 | 7.10 | 7.10 | 7.10 | 7.10 |
| MBC | 14.20 | 14.20 | 7.10 | 14.20 | 14.20 | 14.20 | 14.20 | 14.20 | |
| 5i | MIC | 7.97 | 11.96 | 3.99 | 15.94 | 7.97 | 7.97 | 15.94 | 11.96 |
| MBC | 15.94 | 15.94 | 7.97 | 31.88 | 15.94 | 15.94 | 31.88 | 15.94 | |
| 5j | MIC | 15.37 | 23.05 | 7.68 | 30.74 | 7.68 | 11.53 | 15.37 | 30.74 |
| MBC | 30.74 | 30.74 | 15.37 | 61.48 | 15.37 | 15.37 | 30.74 | 61.48 | |
| 5k | MIC | 3.71 | 7.42 | 3.71 | 7.42 | 7.42 | 5.44 | 7.42 | 11.25 |
| MBC | 7.42 | 14.88 | 7.42 | 14.88 | 14.88 | 7.42 | 14.88 | 14.88 | |
| Amp. | MIC | 24.80 | 24.80 | 24.80 | 37.20 | 24.80 | 74.40 | 24.80 | 37.20 |
| MBC | 37.20 | 37.20 | 37.20 | 74.40 | 37.20 | 124.0 | 49.20 | 49.20 | |
| Strept. | MIC | 4.30 | 8.60 | 17.20 | 25.80 | 4.30 | 17.20 | 17.20 | 17.20 |
| MBC | 8.60 | 17.20 | 34.40 | 51.60 | 8.60 | 34.40 | 34.40 | 34.40 |
| Compounds | Resistant Strains | Biofilm Formation | ||||
|---|---|---|---|---|---|---|
| MRSA | P.a. | E.c. | MIC | 0.5MIC | ||
| 4h | MIC | 0.5 | 0.12 | 0.5 | 17.14 | NE |
| MBC | 1.0 | 0.25 | 1.0 | |||
| 5b | MIC | 0.25 | 0.12 | 0.25 | 37.93 | 22.97 |
| MBC | 0.5 | 0.25 | 0.5 | |||
| 5g | MIC | 0.5 | 0.12 | 0.5 | 30.59 | 11.02 |
| MBC | 1.0 | 0.25 | 1.0 | |||
| Streptomycin | MIC | 0.1 | 0.05 | 0.1 | 71.94 | 55.42 |
| MBC | / | 0.1 | 0.2 | |||
| Ampicilline | MIC | / | 0.2 | 0.2 | 67.36 | 30.35 |
| MBC | / | / | / | |||
| R.br | A.f | A.v | A.o | A.n | T.v | P.o | P.f | Pvc | |
|---|---|---|---|---|---|---|---|---|---|
| 4a | MIC | 18.87 | 18.87 | 9.43 | 18.87 | 6.92 | 6.92 | 9.43 | 18.87 |
| MFC | 44.03 | 37.74 | 18.87 | 37.74 | 9.43 | 18.87 | 18.87 | 37.74 | |
| 4b | MIC | 3.31 | 4.51 | 3.31 | 4.51 | 3.31 | 4.51 | 4.51 | 4.51 |
| MFC | 4.51 | 9.03 | 4.51 | 9.03 | 4.51 | 9.03 | 9.03 | 9.03 | |
| 4c | MIC | 8.61 | 4.31 | 3.16 | 4.31 | 2.30 | 3.16 | 6.31 | 4.31 |
| MFC | 17.22 | 8.61 | 4.31 | 8.61 | 4.31 | 4.31 | 8.61 | 8.61 | |
| 4d | MIC | 2.31 | 4.33 | 2.89 | 4.33 | 2.31 | 4.33 | 4.33 | 2.89 |
| MFC | 4.33 | 8.66 | 4.33 | 8.66 | 4.33 | 8.66 | 8.66 | 4.33 | |
| 4e | MIC | 4.14 | 4.14 | 6.07 | 3.04 | 8.28 | 6.07 | 8.28 | 9.14 |
| MFC | 8.28 | 8.28 | 8.28 | 4.14 | 16.57 | 8.28 | 16.57 | 17.08 | |
| 4f | MIC | 8.32 | 4.16 | 6.10 | 4.16 | 8.32 | 8.32 | 8.32 | 10.12 |
| MFC | 16.64 | 8.32 | 8.32 | 8.32 | 16.64 | 16.64 | 16.64 | 19.76 | |
| 4g | MIC | 3.98 | 3.98 | 2.12 | 3.98 | 2.12 | 3.98 | 3.98 | 3.98 |
| MFC | 7.97 | 7.97 | 3.98 | 7.97 | 3.98 | 7.97 | 7.97 | 7.97 | |
| 4h | MIC | 1.99 | 3.98 | 2.92 | 3.98 | 2.92 | 3.98 | 5.84 | 5.84 |
| MFC | 3.98 | 7.97 | 3.98 | 7.97 | 3.98 | 7.97 | 7.97 | 7.97 | |
| 4i | MIC | 32.04 | 16.02 | 4.00 | 32.04 | 4.00 | 32.04 | 32.04 | 32.04 |
| MFC | 64.09 | 32.04 | 8.01 | 64.09 | 8.01 | 64.09 | 64.09 | 64.09 | |
| 5a | MIC | 18.07 | 4.52 | 9.04 | 9.04 | 4.52 | 4.52 | 9.04 | 9.04 |
| MFC | 36.14 | 9.04 | 18.07 | 18.07 | 9.04 | 9.04 | 18.07 | 18.07 | |
| 5b | MIC | 8.32 | 4.16 | 4.16 | 4.16 | 2.08 | 6.11 | 8.32 | 8.32 |
| MFC | 16.67 | 8.32 | 8.32 | 8.32 | 4.16 | 8.32 | 16.67 | 16.67 | |
| 5c | MIC | 11.46 | 3.82 | 2.80 | 5.61 | 3.82 | 5.61 | 5.61 | 7.64 |
| MFC | 15.29 | 7.64 | 3.82 | 7.64 | 7.64 | 7.64 | 7.64 | 15.29 | |
| 5d | MIC | 29.52 | 29.52 | 1.97 | 5.41 | 3.69 | 10.07 | 5.41 | 14.76 |
| MFC | 59.04 | 59.04 | 3.69 | 7.38 | 7.38 | 14.76 | 7.38 | 29.52 | |
| 5e | MIC | 7.10 | 3.55 | 1.89 | 3.55 | 1.89 | 3.55 | 3.55 | 3.55 |
| MFC | 14.20 | 7.10 | 3.55 | 7.10 | 3.55 | 7.10 | 7.10 | 7.10 | |
| 5f | MIC | 3.67 | 3.67 | 2.69 | 3.67 | 2.69 | 3.67 | 5.39 | 3.67 |
| MFC | 7.34 | 7.34 | 3.67 | 7.34 | 3.67 | 3.67 | 3.67 | 3.67 | |
| 5g | MIC | 7.36 | 5.39 | 3.68 | 3.68 | 3.68 | 3.68 | 7.36 | 3.68 |
| MFC | 14.72 | 7.36 | 7.36 | 7.36 | 7.36 | 7.36 | 14.72 | 7.36 | |
| 5h | MIC | 21.30 | 7.10 | 3.55 | 7.10 | 2.60 | 14.20 | 3.55 | 14.20 |
| MFC | 28.40 | 14.20 | 7.10 | 14.20 | 3.55 | 28.40 | 7.10 | 28.40 | |
| 5i | MIC | 3.99 | 3.99 | 2.13 | 3.99 | 3.99 | 3.99 | 3.99 | 3.99 |
| MFC | 7.97 | 7.97 | 3.99 | 7.97 | 7.97 | 7.97 | 7.97 | 7.97 | |
| 5j | MIC | 7.68 | 3.84 | 2.82 | 3.84 | 2.82 | 3.84 | 3.84 | 5.63 |
| MFC | 15.37 | 7.68 | 3.84 | 7.68 | 3.84 | 7.68 | 7.68 | 7.68 | |
| 5k | MIC | 5.44 | 3.71 | 2.72 | 5.44 | 3.71 | 14.88 | 5.44 | 7.42 |
| MFC | 7.42 | 7.42 | 3.71 | 7.42 | 7.42 | 29.67 | 7.42 | 14.88 | |
| Ketoconazole | MIC | 38.0 | 285.0 | 38.0 | 38.00 | 475.0 | 38.00 | 380.0 | 37.60 |
| MFC | 95.00 | 380.0 | 95.00 | 95.00 | 570.0 | 95.00 | 380.0 | 94.00 | |
| Bifonazole | MIC | 48.00 | 48.0 | 48.00 | 48.00 | 64.00 | 64.00 | 48.00 | 32.20 |
| MFC | 64.00 | 64.0 | 80.00 | 64.00 | 80.00 | 80.00 | 64.00 | 48.30 |
| Est. Binding Energy (kcal/mol) | ||||||
|---|---|---|---|---|---|---|
| Comp. | Gyrase 1KZN | Thymidylate Kinase 4QGG | E. coli MurB 2Q85 | E. coli MurB | 1-H E. coli MurB | Residues E. coli MurB |
| 4a | −1.28 | - | −6.25 | −23.74 | 1 | Arg158 |
| 4b | −6.22 | −2.69 | −9.84 | −30.42 | 2 | Ser228 |
| 4c | −4.36 | - | −7.15 | −26.71 | 2 | Arg158, Arg213 |
| 4d | −4.19 | - | −8.10 | −28.22 | 2 | Arg158, Arg213 |
| 4e | −5.10 | −2.41 | −8.17 | −28.36 | 2 | Ser228, Arg213 |
| 4f | −4.07 | - | −7.11 | −26.55 | 2 | Arg158, Arg213 |
| 4g | −6.25 | −3.14 | −10.08 | −31.16 | 2 | Gly122, Ser228 |
| 4h | −7.09 | −4.66 | −11.25 | −33.49 | 3 | Arg158, Ser228, Asn232 |
| 4i | −5.28 | - | −7.70 | −27.11 | 2 | Tyr189, Ser228 |
| 5a | −3.65 | −3.27 | −6.88 | −24.79 | 2 | Arg158, Tyr189 |
| 5b | −7.15 | −4.19 | −12.33 | −36.27 | 3 | Ser228, Ala226 |
| 5c | −5.13 | −2.26 | −8.25 | −28.41 | 2 | Ser228, Arg213 |
| 5d | −6.92 | −3.27 | −10.51 | −31.44 | 2 | Gly122, Ser228 |
| 5e | −6.20 | - | −9.82 | −30.71 | 2 | Ser228, Ala226 |
| 5f | −4.32 | - | −7.14 | −26.58 | 2 | Arg158, Arg213 |
| 5g | −7.00 | −4.11 | −11.28 | −33.42 | 3 | Arg158, Ser228, Asn232 |
| 5h | −5.87 | −3.15 | −8.75 | −28.98 | 2 | Gly122, Ser228 |
| 5i | - | - | −5.73 | −20.75 | - | - |
| 5j | −2.55 | - | −5.77 | −20.86 | 1 | Arg213 |
| 5k | −5.84 | −3.11 | −8.67 | −28.33 | 2 | Gly122, Ser228 |
| No | Est. Binding Energy (kcal/mol) CYP51 of C. albicans PDB ID: 5V5Z | Binding Affinity Score CYP51 of C. albicans PDB ID: 5V5Z | I-H | Residues CYP51 of C. albicans PDB ID: 5V5Z |
|---|---|---|---|---|
| 17 4a | −3.15 | −15.18 | - | - |
| 4 4b | −8.14 | −27.22 | 1 | Tyr132 |
| 13 4c | −7.15 | −26.02 | 1 | Tyr132 |
| 5 4d | −10.89 | −31.08 | - | HEM601 (ionizable) |
| 14 4e | −4.18 | −15.21 | - | |
| 2 4i | −3.15 | −13.57 | - | - |
| 12 4g | −9.66 | −29.47 | 1 | Tyr132 |
| 15 4h | −10.14 | −30.25 | - | HEM601 (ionizable) |
| 2 4i | −3.15 | −13.57 | - | - |
| 18 5a | −5.12 | −20.96 | - | - |
| 19 5b | −1.14 | −6.29 | - | - |
| 7 5c | −5.16 | −20.85 | 1 | Tyr118 |
| 11 5d | −5.17 | −21.30 | - | - |
| 8 5f | −11.13 | −32.56 | 1 | Tyr132 HEM601 (ionizable, pi) |
| 1 5h | −6.68 | −24.79 | - | - |
| 3 5i | −8.74 | −27.58 | 1 | Tyr64 |
| 9 5j | −8.14 | −26.97 | 1 | Tyr64 |
| 10 5k | −6.25 | −23.88 | 1 | Tyr118 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Horishny, V.; Kartsev, V.; Geronikaki, A.; Matiychuk, V.; Petrou, A.; Glamoclija, J.; Ciric, A.; Sokovic, M. 5-(1H-Indol-3-ylmethylene)-4-oxo-2-thioxothiazolidin-3-yl)alkancarboxylic Acids as Antimicrobial Agents: Synthesis, Biological Evaluation, and Molecular Docking Studies. Molecules 2020, 25, 1964. https://doi.org/10.3390/molecules25081964
Horishny V, Kartsev V, Geronikaki A, Matiychuk V, Petrou A, Glamoclija J, Ciric A, Sokovic M. 5-(1H-Indol-3-ylmethylene)-4-oxo-2-thioxothiazolidin-3-yl)alkancarboxylic Acids as Antimicrobial Agents: Synthesis, Biological Evaluation, and Molecular Docking Studies. Molecules. 2020; 25(8):1964. https://doi.org/10.3390/molecules25081964
Chicago/Turabian StyleHorishny, Volodymyr, Victor Kartsev, Athina Geronikaki, Vasyl Matiychuk, Anthi Petrou, Jasmina Glamoclija, Ana Ciric, and Marina Sokovic. 2020. "5-(1H-Indol-3-ylmethylene)-4-oxo-2-thioxothiazolidin-3-yl)alkancarboxylic Acids as Antimicrobial Agents: Synthesis, Biological Evaluation, and Molecular Docking Studies" Molecules 25, no. 8: 1964. https://doi.org/10.3390/molecules25081964
APA StyleHorishny, V., Kartsev, V., Geronikaki, A., Matiychuk, V., Petrou, A., Glamoclija, J., Ciric, A., & Sokovic, M. (2020). 5-(1H-Indol-3-ylmethylene)-4-oxo-2-thioxothiazolidin-3-yl)alkancarboxylic Acids as Antimicrobial Agents: Synthesis, Biological Evaluation, and Molecular Docking Studies. Molecules, 25(8), 1964. https://doi.org/10.3390/molecules25081964