Evaluation of Enzyme Inhibitory Activity of Flavonoids by Polydopamine-Modified Hollow Fiber-Immobilized Xanthine Oxidase
Abstract
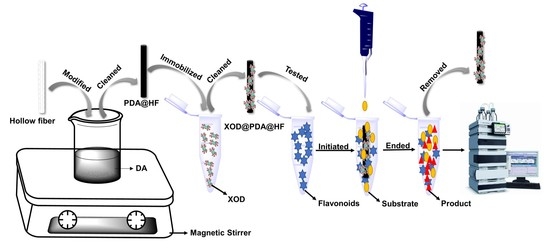
:1. Introduction
2. Results and Discussion
2.1. Characterizations
2.2. Optimization of Preparation Conditions of Hollow Fiber Modified by PDA
2.3. Optimization of XOD Immobilization Conditions
2.4. Effects of pH and Temperature on the Activities of Free and Immobilized XOD
2.5. Performance of the XOD@PDA@HF
2.5.1. Enzyme Kinetics Parameters
2.5.2. Inhibition Kinetics Study
2.5.3. Reproducibility and Reusability
2.6. Inhibitory Activity Evaluation of Flavonoids
2.7. Molecular Docking Study
3. Materials and Methods
3.1. Chemicals and Materials
3.2. Instruments and HPLC-DAD Analysis
3.3. Sample Preparation
3.4. Preparation of XOD@PDA@HF
3.5. XOD Activity Assay
3.6. Characterization of the PDA@HF
3.7. Enzymatic Kinetic Study
3.8. Inhibition Kinetics Study of XOD
3.9. Inhibitory Assay by XOD@PDA@HF
3.10. Molecular Docking Study
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Qi, J.; Sun, L.Q.; Qian, S.Y.; Yu, B.Y. A novel multi-hyphenated analytical method to simultaneously determine xanthine oxidase inhibitors and superoxide anion scavengers in natural products. Anal. Chim. Acta 2017, 984, 124–133. [Google Scholar] [CrossRef]
- Bodade, R.G.; Beedkar, S.D.; Manwar, A.V.; Khobragade, C.N. Homology modeling and docking study of xanthine oxidase of Arthrobacter sp. XL26. Int. J. Biol. Macromol. 2010, 47, 298–303. [Google Scholar] [CrossRef]
- Moradi-Marjaneh, R.; Hassanian, S.M.; Mehramiz, M.; Rezayi, M.; Ferns, G.A.; Khazaei, M.; Avan, A. Reactive oxygen species in colorectal cancer: The therapeutic impact and its potential roles in tumor progression via perturbation of cellular and physiological dysregulated pathways. J. Cell Physiol. 2019, 234, 10072–10079. [Google Scholar] [CrossRef]
- Zhang, H.J.; Hu, Y.J.; Xu, P.; Liang, W.Q.; Zhou, J.; Liu, P.J.; Cheng, L.; Pu, J.B. Screening of Potential Xanthine Oxidase Inhibitors in Gnaphalium hypoleucum DC. by Immobilized Metal Affinity Chromatography and Ultrafiltration-Ultra Performance Liquid Chromatography-Mass Spectrometry. Molecules 2016, 21, 1242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Day, R.O.; Graham, G.G.; Hicks, M.; McLachlan, A.J.; Stocker, S.L.; Williams, K.M. Clinical pharmacokinetics and pharmacodynamics of allopurinol and oxypurinol. Clin. Pharmacokinet. 2007, 46, 623–644. [Google Scholar] [CrossRef] [PubMed]
- Ishii, T.; Taguri, M.; Tamura, K.; Oyama, K. Evaluation of the Effectiveness of Xanthine Oxidoreductase Inhibitors on Haemodialysis Patients using a Marginal Structural Model. Sci. Rep. 2017, 7, 14004. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ninfali, P.; Antonini, E.; Frati, A.; Scarpa, E.S. C-Glycosyl Flavonoids from Beta vulgaris Cicla and Betalains from Beta vulgaris rubra: Antioxidant, Anticancer and Antiinflammatory Activities-A Review. Phytother. Res. 2017, 31, 871–884. [Google Scholar] [CrossRef] [PubMed]
- López, J.G. Flavonoids in Health and Disease. Curr. Med. Chem. 2019, 26, 6972–6975. [Google Scholar] [CrossRef]
- Owen, P.L.; Johns, T. Xanthine oxidase inhibitory activity of northeastern North American plant remedies used for gout. J. Ethnopharmacol. 1999, 64, 149–160. [Google Scholar] [CrossRef]
- Chen, C.H.; Chan, H.C.; Chu, Y.T.; Ho, H.Y.; Chen, P.Y.; Lee, T.H.; Lee, C.K. Antioxidant activity of some plant extracts towards xanthine oxidase, lipoxygenase and tyrosinase. Molecules 2009, 14, 2947–2958. [Google Scholar] [CrossRef]
- Santi, M.D.; Zunini, M.P.; Vera, B.; Bouzidi, C.; Dumontet, V.; Abin-Carriquiry, A.; Grougnet, R.; Ortega, M.G. Xanthine oxidase inhibitory activity of natural and hemisynthetic flavonoids from Gardenia oudiepe (Rubiaceae) in vitro and molecular docking studies. Eur. J. Med. Chem. 2018, 143, 577–582. [Google Scholar] [CrossRef] [PubMed]
- Wahab, R.A.; Elias, N.; Abdullah, F.; Ghoshal, S.K. On the taught new tricks of enzymes immobilization: An all-inclusive overview. React. Funct. Polym. 2020, 152, 104613. [Google Scholar] [CrossRef]
- Zhang, H.; Lu, M.; Jiang, H.; Wu, Z.Y.; Zhou, D.D.; Li, D.Q.; Yang, F.Q. Tyrosinase-mediated dopamine polymerization modified magnetic alginate beads for dual-enzymes encapsulation: Preparation, performance and application. Colloids Surf. B Biointerfaces 2020, 188, 110800. [Google Scholar] [CrossRef]
- Mohamad, N.R.; Marzuki, N.H.; Buang, N.A.; Huyop, F.; Wahab, R.A. An overview of technologies for immobilization of enzymes and surface analysis techniques for immobilized enzymes. Biotechnol. Biotechnol. Equip. 2015, 29, 205–220. [Google Scholar] [CrossRef] [PubMed]
- Datta, S.; Christena, L.R.; Rajaram, Y.R. Enzyme immobilization: An overview on techniques and support materials. 3 Biotech. 2013, 3, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.J.; Caruso, F. Mesoporous silica spheres as supports for enzyme immobilization and encapsulation. Chem. Mat. 2005, 17, 953–961. [Google Scholar] [CrossRef]
- Sassolas, A.; Blum, L.J.; Leca-Bouvier, B.D. Immobilization strategies to develop enzymatic biosensors. Biotechnol. Adv. 2012, 30, 489–511. [Google Scholar] [CrossRef]
- Zhang, H.; Wu, Z.Y.; Yang, Y.Y.; Yang, F.Q.; Li, S.P. Recent applications of immobilized biomaterials in herbal analysis. J. Chromatogr. A. 2019, 1603, 216–230. [Google Scholar] [CrossRef]
- Zhang, H.; Lu, M.; Jiang, H.; Wang, X.; Yang, F.Q. Evaluation inhibitory activity of catechins on trypsin by capillary electrophoresis-based immobilized enzyme microreactor with chromogenic substrate. J. Sep. Sci. 2020, 43, 3136–3145. [Google Scholar] [CrossRef]
- Tao, Y.; Zhang, Y.; Wang, Y.; Cheng, Y. Hollow fiber based affinity selection combined with high performance liquid chromatography-mass spectroscopy for rapid screening lipase inhibitors from lotus leaf. Anal. Chim. Acta. 2013, 785, 75–81. [Google Scholar] [CrossRef]
- Zhao, C.P.; Yin, S.J.; Chen, G.Y.; Wang, Y.; Chen, H.; Zhao, J.; Yang, F.Q. Adsorbed hollow fiber immobilized tyrosinase for the screening of enzyme inhibitors from Pueraria lobata extract. J. Pharm. Biomed. Anal. 2021, 193, 113743. [Google Scholar] [CrossRef]
- Lee, H.; Dellatore, S.M.; Miller, W.M.; Messersmith, P.B. Mussel-inspired surface chemistry for multifunctional coatings. Science 2007, 318, 426–430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, J.; Yang, C.; Zhang, S.; Wang, X.; Jiang, Z.; Zhang, W.; Song, X.; Ai, Q.; Tian, C. Polydopamine microcapsules with different wall structures prepared by a template-mediated method for enzyme immobilization. ACS. Appl. Mater. Interfaces 2013, 5, 9991–9997. [Google Scholar] [CrossRef]
- Li, D.P.; Xu, L.; Qi, J.; Yu, B.Y. Screening and analysis of cyclooxygenase-2 inhibitors from the complex matrix: A case study to illustrate the important effect of immobilized enzyme activity in magnetic ligand fishing. J. Pharm. Biomed. Anal. 2019, 175, 112795. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.Y.; Zhang, H.; Li, F.; Yang, F.Q. Evaluation of xanthine oxidase inhibitory activity of flavonoids by an online capillary electrophoresis-based immobilized enzyme microreactor. Electrophoresis 2020, 41, 1326–1332. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, M.V.N.; Rodrigues-Silva, C.; Boaventura, S.; Oliveira, A.S.S.; Rath, S.; Cass, Q.B. On-Flow LC-MS/MS method for screening of xanthine oxidase inhibitors. J. Pharm. Biomed. Anal. 2020, 181, 113097. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Li, D.; Yu, B.; Qi, J. Screening inhibitors of xanthine oxidase from natural products using enzyme immobilized magnetic beads by high-performance liquid chromatography coupled with tandem mass spectrometry. J. Sep. Sci. 2017, 40, 1877–1886. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Chen, Z. Screening of neuraminidase inhibitors from traditional Chinese medicines by integrating capillary electrophoresis with immobilized enzyme microreactor. J. Chromatogr. A 2014, 1340, 139–145. [Google Scholar] [CrossRef]
- Lin, W.Q.; Xie, J.X.; Wu, X.M.; Yang, L.; Wang, H.D. Inhibition of xanthine oxidase activity by gnaphalium affine extract. Chin. Med. Sci. J. 2014, 29, 225–230. [Google Scholar] [CrossRef]
- Lu, J.; Song, H.P.; Li, P.; Zhou, P.; Dong, X.; Chen, J. Screening of direct thrombin inhibitors from Radix Salviae Miltiorrhizae by a peak fractionation approach. J. Pharm. Biomed. Anal. 2015, 109, 85–90. [Google Scholar] [CrossRef] [PubMed]
- Sorouraddin, M.H.; Amini, K.; Naseri, A.; Vallipour, J.; Hanaee, J.; Rashidi, M.R. A new multi-wavelength model-based method for determination of enzyme kinetic parameters. J. Biosci. 2010, 35, 395–403. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.Y.; Zhang, H.; Yang, F.Q. A simple and portable method for β-Glucosidase activity assay and its inhibitor screening based on a personal glucose meter. Anal. Chim. Acta 2021, 1142, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Prusoff, W.H. Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem. Pharmacol. 1973, 22, 3099–3108. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [Green Version]
- Pauff, J.M.; Cao, H.; Hille, R. Substrate Orientation and Catalysis at the Molybdenum Site in Xanthine Oxidase: Crystal structures in complex with xanthine and lumazine. J. Biol. Chem. 2009, 284, 8760–8767. [Google Scholar] [CrossRef] [Green Version]
- Banner, D.W.; Hadváry, P. Crystallographic analysis at 3.0-A resolution of the binding to human thrombin of four active site-directed inhibitors. J. Biol. Chem. 1991, 266, 20085–20093. [Google Scholar] [CrossRef]








| Compounds | % of Inhibition | Compounds | % of Inhibition |
|---|---|---|---|
| Quercetin | 79.86 ± 3.50 | Epicatechin | 16.61 ± 3.03 |
| Apigenin | 80.98 ± 0.64 | Epigallocatechin | 54.92 ± 0.41 |
| Puerarin | 61.15 ± 6.26 | Epicatechin gallate | 26.80 ± 5.78 |
| Catechin | 28.68 ± 0.60 | Epigallocatechin gallate | 33.07 ± 3.39 |
| Compounds | Binding Energy (Kcal/mol) | Amino Acid Residues | Hydrogen Bonds |
|---|---|---|---|
| Allopurinol | −5.68 | GLY796, MET1038, GLY797, GLN1040, CYS150, PHE798, ARG912, GLN1194 | GLY797, GLN1194, CYS150, MET1038 |
| Quercetin | −5.81 | ALA338, GLY339, GLY46, GLY47, ARG426, SER1225, LYS1228, ALA1231, ILE1229, LEU147 | GLY46, GLY47, ARG426, LYS1228, ILE1229 |
| Apigenin | −6.74 | GLU45, GLY47, LEU147, ALA338, TYR1227, LYS1228 | GLU45, TYR1227 |
| Puerarin | −6.54 | GLN144, TRP336, GLY1233, SER1234, ARG426, ALA424, LYS1228 | GLN144, SER1234, GLY1233 |
| Epigallocatechin | −6.98 | GLN144, TRP336, ALA1231, LEU147, GLY47, ILE1229, LYS1228, TYR1227 | TYR1227, GLY47, GLN144, ILE1229, TRP336 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, C.-P.; Chen, G.-Y.; Wang, Y.; Chen, H.; Yu, J.-W.; Yang, F.-Q. Evaluation of Enzyme Inhibitory Activity of Flavonoids by Polydopamine-Modified Hollow Fiber-Immobilized Xanthine Oxidase. Molecules 2021, 26, 3931. https://doi.org/10.3390/molecules26133931
Zhao C-P, Chen G-Y, Wang Y, Chen H, Yu J-W, Yang F-Q. Evaluation of Enzyme Inhibitory Activity of Flavonoids by Polydopamine-Modified Hollow Fiber-Immobilized Xanthine Oxidase. Molecules. 2021; 26(13):3931. https://doi.org/10.3390/molecules26133931
Chicago/Turabian StyleZhao, Cong-Peng, Guo-Ying Chen, Yuan Wang, Hua Chen, Jia-Wen Yu, and Feng-Qing Yang. 2021. "Evaluation of Enzyme Inhibitory Activity of Flavonoids by Polydopamine-Modified Hollow Fiber-Immobilized Xanthine Oxidase" Molecules 26, no. 13: 3931. https://doi.org/10.3390/molecules26133931
APA StyleZhao, C. -P., Chen, G. -Y., Wang, Y., Chen, H., Yu, J. -W., & Yang, F. -Q. (2021). Evaluation of Enzyme Inhibitory Activity of Flavonoids by Polydopamine-Modified Hollow Fiber-Immobilized Xanthine Oxidase. Molecules, 26(13), 3931. https://doi.org/10.3390/molecules26133931