Reconstitution of Caveolin-1 into Artificial Lipid Membrane: Characterization by Transmission Electron Microscopy and Solid-State Nuclear Magnetic Resonance
Abstract
:1. Introduction
2. Results
2.1. Protein Production
2.2. Protein Oligomerization
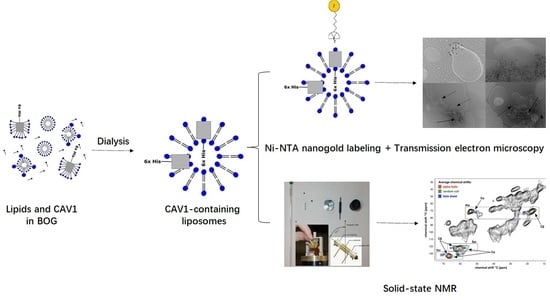
2.3. Membrane Reconstitution
2.4. ssNMR Analysis of CAV1
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Plasmid Preparation
4.3. Protein Expression
4.4. Protein Purification
4.5. Protein Characterization
4.6. In-Gel Tryptic Digestion
4.7. Membrane Reconstitution
4.8. TEM
4.9. CD Spectroscopy
4.10. Peptide Synthesis and Purification
4.11. ThT Binding Assay
4.12. ssNMR Spectroscopy
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Parton, R.G.; Tillu, V.A.; Collins, B.M. Caveolae. Curr. Biol. 2018, 28, 402–405. [Google Scholar] [CrossRef] [Green Version]
- Parton, R.G.; McMahon, K.A.; Wu, Y. Caveolae: Formation, Dynamics, and Function. Curr. Opin. Cell Biol. 2020, 65, 8–16. [Google Scholar] [CrossRef]
- Parton, R.G.; del Pozo, M.A.; Vassilopoulos, S.; Nabi, I.R.; Le Lay, S.; Lundmark, R.; Kenworthy, A.K.; Camus, A.; Blouin, C.M.; Sessa, W.C.; et al. Caveolae: The FAQs. Traffic 2020, 21, 181–185. [Google Scholar] [CrossRef]
- Yamada, E. The Fine Structure of the Gall Bladder Epithelium of the Mouse. J. Biophys. Biochem. Cytol. 1955, 1, 445–458. [Google Scholar] [CrossRef] [Green Version]
- Del Pozo, M.A.; Lolo, F.N.; Echarri, A. Caveolae: Mechanosensing and Mechanotransduction Devices Linking Membrane Trafficking to Mechanoadaptation. Curr. Opin. Cell Biol. 2021, 68, 113–123. [Google Scholar] [CrossRef]
- Filippini, A.; D’alessio, A. Caveolae and Lipid Rafts in Endothelium: Valuable Organelles for Multiple Functions. Biomolecules 2020, 10, 1218. [Google Scholar] [CrossRef] [PubMed]
- Parton, R.G.; Kozlov, M.M.; Ariotti, N. Caveolae and Lipid Sorting: Shaping the Cellular Response to Stress. J. Cell Biol. 2020, 219, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Wang, J.; Hou, W.; Shan, Y.; Wang, S.; Liu, F. Dynamic Dissection of the Endocytosis of Porcine Epidemic Diarrhea Coronavirus Cooperatively Mediated by Clathrin and Caveolae as Visualized by Single-Virus Tracking. MBio 2021, 12, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Ma, J.; Wang, Z.; Li, H.; Shen, H.; Li, X.; Chen, G. Mfsd2a Attenuates Blood-Brain Barrier Disruption after Sub-Arachnoid Hemorrhage by Inhibiting Caveolae-Mediated Transcellular Transport in Rats. Transl. Stroke Res. 2020, 11, 1012–1027. [Google Scholar] [CrossRef]
- Murata, M.; Peränen, J.; Schreiner, R.; Wieland, F.; Kurzchalia, T.V.; Simons, K. VIP21/Caveolin is a Cholesterol-Binding Protein. Proc. Natl. Acad. Sci. USA 1995, 92, 10339–10343. [Google Scholar] [CrossRef] [Green Version]
- Rothberg, K.G.; Heuser, J.E.; Donzell, W.C.; Ying, Y.-S.; Glenney, J.R.; Anderson, R.G.W. Caveolin, a Protein Component of Caveolae Membrane Coats. Cell 1992, 68, 673–682. [Google Scholar] [CrossRef]
- Echarri, A.; Del Pozo, M.A. Caveolae. Curr. Biol. 2012, 22, 114–116. [Google Scholar] [CrossRef] [Green Version]
- Yoon, H.J.; Kim, D.H.; Kim, S.J.; Jang, J.H.; Surh, Y.J. Src-Mediated Phosphorylation, Ubiquitination and Degradation of Caveolin-1 Promotes Breast Cancer Cell Stemness. Cancer Lett. 2019, 449, 8–19. [Google Scholar] [CrossRef]
- Hu, L.; Xu, X.; Li, Q.; Chen, X.; Yuan, X.; Qiu, S.; Yao, C.; Zhang, D.; Wang, F. Caveolin-1 Increases Glycolysis in Pancreatic Cancer Cells and Triggers Cachectic States. FASEB J. 2021, 35, e21826. [Google Scholar] [CrossRef]
- Vykoukal, J.; Fahrmann, J.F.; Gregg, J.R.; Tang, Z.; Basourakos, S.; Irajizad, E.; Park, S.; Yang, G.; Creighton, C.J.; Fleury, A.; et al. Caveolin-1-Mediated Sphingolipid Oncometabolism Underlies a Metabolic Vulnerability of Prostate Cancer. Nat. Commun. 2020, 11, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.; Li, Y.; Li, Y.; Wang, Q. Caveolin-1, a Novel Player in Cognitive Decline. Neurosci. Biobehav. Rev. 2021, 129, 95–106. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Jiang, W.; Xin, J.; Xue, W.; Shi, C.; Wen, J.; Huang, Y.; Hu, C. Caveolin-1 Alleviates Acetaminophen-Induced Fat Accumulation in Non-Alcoholic Fatty Liver Disease by Enhancing Hepatic Antioxidant Ability via Activating AMPK Pathway. Front. Pharmacol. 2021, 12, 1–16. [Google Scholar] [CrossRef]
- Fujimoto, T.; Kogo, H.; Nomura, R.; Une, T. Isoforms of Caveolin-1 and Caveolar Structure. J. Cell Sci. 2000, 113, 3509–3517. [Google Scholar] [CrossRef]
- Volonte, D.; McTiernan, C.F.; Drab, M.; Kasper, M.; Galbiati, F. Caveolin-1 and Caveolin-3 Form Heterooligomeric Complexes in Atrial Cardiac Myocytes That Are Required for Doxorubicin-Induced Apoptosis. Am. J. Physiol.-Heart Circ. Physiol. 2008, 294, 392–401. [Google Scholar] [CrossRef] [Green Version]
- Scherer, P.E.; Lewis, R.Y.; Volonté, D.; Engelman, J.A.; Galbiati, F.; Couet, J.; Kohtz, D.S.; Van Donselaar, E.; Peters, P.; Lisanti, M.P. Cell-Type and Tissue-Specific Expression of Caveolin-2. Caveolins 1 and 2 Co-Localize and Form a Stable Hetero-Oligomeric Complex In Vivo. J. Biol. Chem. 1997, 272, 29337–29346. [Google Scholar] [CrossRef] [Green Version]
- Rieth, M.D.; Lee, J.; Glover, K.J. Probing the Caveolin-1 P132L Mutant: Critical Insights into Its Oligomeric Behavior and Structure. Biochemistry 2012, 51, 3911–3918. [Google Scholar] [CrossRef] [Green Version]
- Khater, I.M.; Liu, Q.; Chou, K.C.; Hamarneh, G.; Nabi, I.R. Super-Resolution Modularity Analysis Shows Polyhedral Caveolin-1 Oligomers Combine to Form Scaffolds and Caveolae. Sci. Rep. 2019, 9, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Han, B.; Porta, J.C.; Hanks, J.L.; Peskova, Y.; Binshtein, E.; Dryden, K.; Claxton, D.P.; McHaourab, H.S.; Karakas, E.; Ohi, M.D.; et al. Structure and Assembly of CAV1 8S Complexes Revealed by Single Particle Electron Microscopy. Sci. Adv. 2020, 6, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Glover, K.J.; Im, W. U-Shaped Caveolin-1 Conformations Are Tightly Regulated by Hydrogen Bonds with Lipids. J. Comput. Chem. 2019, 40, 1570–1577. [Google Scholar] [CrossRef] [PubMed]
- Root, K.T.; Glover, K.J. Reconstitution and Spectroscopic Analysis of Caveolin-1 Residues 62-178 Reveals that Proline 110 Governs Its Structure and Solvent Exposure. Biochim. Biophys. Acta-Biomembr. 2016, 1858, 682–688. [Google Scholar] [CrossRef] [PubMed]
- Krishna, A.; Sengupta, D. Interplay between Membrane Curvature and Cholesterol: Role of Palmitoylated Caveolin-1. Biophys. J. 2019, 116, 69–78. [Google Scholar] [CrossRef] [Green Version]
- Hoop, C.L.; Sivanandam, V.N.; Kodali, R.; Srnec, M.N.; Van Der Wel, P.C.A. Structural Characterization of the Caveolin Scaffolding Domain in Association with Cholesterol-Rich Membranes. Biochemistry 2012, 51, 90–99. [Google Scholar] [CrossRef]
- Root, K.T.; Plucinsky, S.M.; Glover, K.J. Recent Progress in the Topology, Structure, and Oligomerization of Caveolin: A Building Block of Caveolae; Elsevier Ltd.: Amsterdam, The Netherlands, 2015; Volume 75. [Google Scholar]
- Plucinsky, S.M.; Glover, K.J. Secondary Structure Analysis of a Functional Construct of Caveolin-1 Reveals a Long C-Terminal Helix. Biophys. J. 2015, 109, 1686–1688. [Google Scholar] [CrossRef] [Green Version]
- Smith, J.N.; Edgar, J.M.; Balk, J.M.; Iftikhar, M.; Fong, J.C.; Olsen, T.J.; Fishman, D.A.; Majumdar, S.; Weiss, G.A. Directed Evolution and Biophysical Characterization of a Full-Length, Soluble, Human Caveolin-1 Variant. Biochim. Biophys. Acta-Proteins Proteom. 2018, 1866, 963–972. [Google Scholar] [CrossRef]
- Williams, J.K.; Tietze, D.; Lee, M.; Wang, J.; Hong, M. Solid-State NMR Investigation of the Conformation, Proton Conduction, and Hydration of the Influenza B Virus M2 Transmembrane Proton Channel. J. Am. Chem. Soc. 2016, 138, 8143–8155. [Google Scholar] [CrossRef] [Green Version]
- Wang, T.; Park, Y.B.; Caporini, M.A.; Rosay, M.; Zhong, L.; Cosgrove, D.J.; Hong, M. Sensitivity-Enhanced Solid-State NMR Detection of Expansin’s Target in Plant Cell Walls. Proc. Natl. Acad. Sci. USA 2013, 110, 16444–16449. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luca, S.; Yau, W.-M.; Leapman, R.; Tycko, R. Peptide Conformation and Supramolecular Organization in Amylin Fibrils: Constraints from Solid-State NMR. Biochemistry 2007, 46, 13505–13522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernandez, I.; Ying, Y.; Albanesi, J.; Anderson, R.G.W. Mechanism of Caveolin Filament Assembly. Proc. Natl. Acad. Sci. USA 2002, 99, 11193–11198. [Google Scholar] [CrossRef] [Green Version]
- Sargiacomo, M.; Scherer, P.E.; Tang, Z.L.; Kübler, E.; Song, K.S.; Sanders, M.C.; Lisanti, M.P. Oligomeric Structure of Caveolin: Implications for Caveolae Membrane Organization. Proc. Natl. Acad. Sci. USA 1995, 92, 9407–9411. [Google Scholar] [CrossRef] [Green Version]
- Rovnyagina, N.R.; Budylin, G.S.; Vainer, Y.G.; Tikhonova, T.N.; Vasin, S.L.; Yakovlev, A.A.; Kompanets, V.O.; Chekalin, S.V.; Priezzhev, A.V.; Shirshin, E.A. Fluorescence Lifetime and Intensity of Thioflavin T as Reporters of Different Fibrillation Stages: Insights Obtained from Fluorescence Up-Conversion and Particle Size Distribution Measurements. Int. J. Mol. Sci. 2020, 21, 6169. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Aleksandrov, L.A.; Riordan, J.R.; Ford, R.C. Do Main Location within the Cystic Fibrosis Transmembrane Conductance Regulator Protein Investigated by Electron Microscopy and Gold Labelling. Biochim. Biophys. Acta-Biomembr. 2011, 1808, 399–404. [Google Scholar] [CrossRef]
- Zhang, X.; Bayles, K.W.; Luca, S. Staphylococcus aureus CidC Is a Pyruvate: Menaquinone Oxidoreductase. Biochemistry 2017, 56, 4819–4829. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ranjit, D.K.; Endres, J.L.; Bayles, K.W. Staphylococcus aureus CidA and LrgA Proteins Exhibit Holin-Like Properties. J. Bacteriol. 2011, 193, 2468–2476. [Google Scholar] [CrossRef] [Green Version]
- Song, K.S.; Li, S.; Okamoto, T.; Quilliam, L.A.; Sargiacomo, M.; Lisanti, M.P. Co-Purification and Direct Interaction of Ras with Caveolin, an Integral Membrane Protein of Caveolae Microdomains: Detergent-Free Purification of Caveolae Membranes. J. Biol. Chem. 1996, 271, 9690–9697. [Google Scholar] [CrossRef] [Green Version]
- Sengupta, D. Cholesterol Modulates the Structure, Binding Modes, and Energetics of Caveolin-Membrane Interactions. J. Phys. Chem. B 2012, 116, 14556–14564. [Google Scholar] [CrossRef] [PubMed]
- Edidin, M. The State of Lipid Rafts: From Model Membranes to Cells. Annu. Rev. Biophys. Biomol. Struct. 2003, 32, 257–283. [Google Scholar] [CrossRef] [Green Version]
- Brown, D.A.; Rose, J.K. Sorting of GPI-Anchored Proteins to Glycolipid-Enriched Membrane Subdomains during Transport to the Apical Cell Surface. Cell 1992, 68, 533–544. [Google Scholar] [CrossRef]
- Pike, L.J.; Han, X.; Chung, K.-N.; Gross, R.W. Lipid Rafts Are Enriched in Arachidonic Acid and Plasmenylethanolamine and Their Composition Is Independent of Caveolin-1 Expression: A Quantitative Electrospray Ionization/Mass Spectrometric Analysis. Biochemistry 2002, 41, 2075–2088. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Sykes, B.D. The 13C Chemical-Shift Index: A Simple Method for the Identification of Protein Secondary Structure Using 13C Chemical-Shift Data. J. Biomol. NMR 1994, 4, 171–180. [Google Scholar] [CrossRef]
- Wang, Y. Probability-Based Protein Secondary Structure Identification Using Combined NMR Chemical-Shift Data. Protein Sci. 2002, 11, 852–861. [Google Scholar] [CrossRef]
- Lige, B.; Romano, J.D.; Sampels, V.; Sonda, S.; Joiner, K.A.; Coppens, I. Introduction of Caveolae Structural Proteins into the Protozoan Toxoplasma Results in the Formation of Heterologous Caveolae but Not Caveolar Endocytosis. PLoS ONE 2012, 7, 1–23. [Google Scholar] [CrossRef] [Green Version]
- Ariotti, N.; Rae, J.; Leneva, N.; Ferguson, C.; Loo, D.; Okano, S.; Hill, M.M.; Walser, P.; Collins, B.M.; Parton, R.G. Molecular Characterization of Caveolin-Induced Membrane Curvature. J. Biol. Chem. 2015, 290, 24875–24890. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jung, W.R.; Sierecki, E.; Bastiani, M.; O’Carroll, A.; Alexandrov, K.; Rae, J.; Johnston, W.; Hunter, D.J.B.; Ferguson, C.; Gambin, Y.; et al. Cell-Free Formation and Interactome Analysis of Caveolae. J. Cell Biol. 2018, 217, 2141–2165. [Google Scholar] [CrossRef]
- Rieth, M.D.; Root, K.T.; Glover, K.J. Reconstitution of Full-Length Human Caveolin-1 into Phospholipid Bicelles: Validation by Analytical Ultracentrifugation. Biophys. Chem. 2020, 259, 106339. [Google Scholar] [CrossRef] [PubMed]
- Walser, P.J.; Ariotti, N.; Howes, M.; Ferguson, C.; Webb, R.; Schwudke, D.; Leneva, N.; Cho, K.J.; Cooper, L.; Rae, J.; et al. Constitutive Formation of Caveolae in a Bacterium. Cell 2012, 150, 752–763. [Google Scholar] [CrossRef] [Green Version]
- Song, K.S.; Tang, Z.; Li, S.; Lisanti, M.P. Mutational Analysis of the Properties of Caveolin-1: A Novel Role for the C-Terminal Domain in Mediating Homo-Typic Caveolin-Caveolin Interactions. J. Biol. Chem. 1997, 272, 4398–4403. [Google Scholar] [CrossRef] [Green Version]
- Inaba, T.; Kishimoto, T.; Murate, M.; Tajima, T.; Sakai, S.; Abe, M.; Makino, A.; Tomishige, N.; Ishitsuka, R.; Ikeda, Y.; et al. Phospholipase Cβ1 Induces Membrane Tubulation and is Involved in Caveolae Formation. Proc. Natl. Acad. Sci. USA 2016, 113, 7834–7839. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hansen, C.G.; Shvets, E.; Howard, G.; Riento, K.; Nichols, B.J. Deletion of Cavin Genes Reveals Tissue-Specific Mechanisms for Morphogenesis of Endothelial Caveolae. Nat. Commun. 2013, 4, 1831. [Google Scholar] [CrossRef] [Green Version]
- Castellani, F.; van Rossum, B.; Diehl, A.; Schubert, M.; Rehbein, K.; Oschkinat, H. Structure of a Protein Determined by Solid-State Magic-Angle-Spinning NMR Spectroscopy. Nature 2002, 420, 99–102. [Google Scholar] [CrossRef] [PubMed]
- Hong, M.; Jakes, K. Selective and Extensive 13C Labeling of a Membrane Protein for Solid- State NMR Investigations. J. Biomol. NMR 1999, 14, 71–74. [Google Scholar] [CrossRef] [PubMed]
- Opella, S.J. Structure Determination of Membrane Proteins in Their Native Phospholipid Bilayer Environment by Rotationally Aligned Solid-State NMR Spectroscopy. Acc. Chem. Res. 2013, 46, 2145–2153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okada, S.; Raja, S.A.; Okerblom, J.; Boddu, A.; Horikawa, Y.; Ray, S.; Okada, H.; Kawamura, I.; Murofushi, Y.; Murray, F.; et al. Deletion of Caveolin Scaffolding Domain Alters Cancer Cell Migration. Cell Cycle 2019, 18, 1268–1280. [Google Scholar] [CrossRef]
- Weng, P.; Zhang, X.T.; Sheng, Q.; Tian, W.F.; Chen, J.L.; Yuan, J.J.; Zhang, J.R.; Pang, Q.F. Caveolin-1 Scaffolding Domain Peptides Enhance Antiinflammatory Effect of Heme Oxygenase-1 through Interrupting Its Interact with Caveolin-1. Oncotarget 2017, 8, 40104–40114. [Google Scholar] [CrossRef] [Green Version]
- Lu, J.; Zhang, J.; Wang, Y.; Sun, Q. Caveolin-1 Scaffolding Domain Peptides Alleviate Liver Fibrosis by Inhibiting TGF-β1/Smad Signaling in Mice. Int. J. Mol. Sci. 2018, 19, 1729. [Google Scholar] [CrossRef] [Green Version]
- Gopu, V.; Fan, L.; Shetty, R.S.; Nagaraja, M.R.; Shetty, S. Caveolin-1 Scaffolding Domain Peptide Regulates Glucose Metabolism in Lung Fibrosis. JCI Insight 2020, 5, 1–17. [Google Scholar] [CrossRef]
- Cai, M.; Huang, Y.; Craigie, R.; Clore, G.M. A Simple Protocol for Expression of Isotope-Labeled Proteins in Escherichia coli Grown in Shaker Flasks at High Cell Density. J. Biomol. NMR 2019, 73, 743–748. [Google Scholar] [CrossRef] [PubMed]
- Duong, N.T.; Lee, D.; Mentink-Vigier, F.; Lafon, O.; De Paëpe, G. On the use of Radio-Frequency Offsets for Improving Double-Quantum Homonuclear Dipolar Recoupling of Half-Integer-Spin Quadrupolar Nuclei. Magn. Reson. Chem. 2021, 59, 991–1008. [Google Scholar] [CrossRef] [PubMed]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Zhang, X.; Kong, W.; Wang, S. Reconstitution of Caveolin-1 into Artificial Lipid Membrane: Characterization by Transmission Electron Microscopy and Solid-State Nuclear Magnetic Resonance. Molecules 2021, 26, 6201. https://doi.org/10.3390/molecules26206201
Zhang Y, Zhang X, Kong W, Wang S. Reconstitution of Caveolin-1 into Artificial Lipid Membrane: Characterization by Transmission Electron Microscopy and Solid-State Nuclear Magnetic Resonance. Molecules. 2021; 26(20):6201. https://doi.org/10.3390/molecules26206201
Chicago/Turabian StyleZhang, Yanli, Xinyan Zhang, Wenru Kong, and Shuqi Wang. 2021. "Reconstitution of Caveolin-1 into Artificial Lipid Membrane: Characterization by Transmission Electron Microscopy and Solid-State Nuclear Magnetic Resonance" Molecules 26, no. 20: 6201. https://doi.org/10.3390/molecules26206201
APA StyleZhang, Y., Zhang, X., Kong, W., & Wang, S. (2021). Reconstitution of Caveolin-1 into Artificial Lipid Membrane: Characterization by Transmission Electron Microscopy and Solid-State Nuclear Magnetic Resonance. Molecules, 26(20), 6201. https://doi.org/10.3390/molecules26206201