Validation of a Fast and Simple HPLC-UV Method for the Quantification of Adenosine Phosphates in Human Bronchial Epithelial Cells
Abstract
:1. Introduction
2. Results
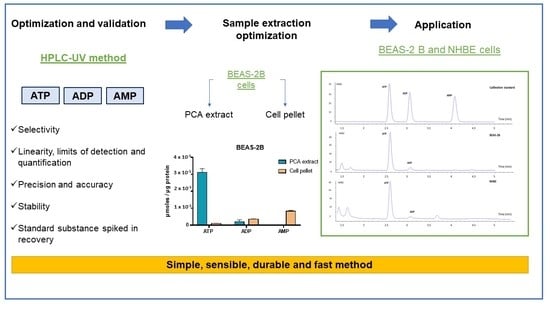
2.1. Method Optimization
2.2. Method Validation
2.2.1. Selectivity
2.2.2. Linearity, the Limits of Detection and Quantification
2.2.3. Precision and Accuracy
2.2.4. Stability
2.2.5. Standard Substance Spiked in Recovery
2.3. Extraction Optimization
2.4. Application to a 3D Bronchial Epithelial Cell Model (NHBE)
3. Discussion
4. Materials and Methods
4.1. Chemicals and Reagents
4.2. Instrumentation and Chromatographic Conditions
4.3. Standard Solution Preparation
4.4. Cell Culture and Sample Extraction
4.4.1. Cell Culture
4.4.2. Sample Extraction
4.5. Method Validation
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Erecińska, M.; Silver, I.A. ATP and Brain Function. Br. J. Pharmacol. 1989, 9, 2–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, T.; Sun, G.; Sun, X.; Zhao, L.; Zhong, R.; Peng, Y. Tumor Energy Metabolism and Potential of 3-Bromopyruvate as an Inhibitor of Aerobic Glycolysis: Implications in Tumor Treatment. Cancers 2019, 11, 317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garland, J.M.; Halestrap, A. Energy Metabolism during Apoptosis. J. Biol. Chem. 1997, 272, 4680–4688. [Google Scholar] [CrossRef] [Green Version]
- Gu, X.; Yan, Y.; Novick, S.J.; Kovach, A.; Goswami, D.; Ke, J.; Tan, M.H.E.; Wang, L.; Li, X.; de Waal, P.; et al. Deconvoluting AMP-activated protein kinase (AMPK) adenine nucleotide binding and sensing. J. Biol. Chem. 2017, 292, 12653–12666. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gorman, M.W.; Feigl, E.O.; Buffington, C.W. Human Plasma ATP Concentration. Clin. Chem. 2007, 53, 318–325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yegutkin, G.G. Nucleotide- and Nucleoside-Converting Ectoenzymes: Important Modulators of Purinergic Signalling Cascade. Biochim. Biophys. Acta 2008, 1783, 673–694. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khlyntseva, S.V.; Bazel’, Y.R.; Vishnikin, A.B.; Andruch, V. Methods for the determination of adenosine triphosphate and other adenine nucleotides. J. Anal. Chem. 2009, 64, 657–673. [Google Scholar] [CrossRef]
- Gu, X.; Ma, Y.; Liu, Y.; Wan, Q. Measurement of mitochondrial respiration in adherent cells by Seahorse XF96 Cell Mito Stress Test. STAR Protoc. 2020, 2, 100245. [Google Scholar] [CrossRef] [PubMed]
- Menegollo, M.; Tessari, I.; Bubacco, L.; Szabadkai, G. Determination of ATP, ADP, and AMP Levels by Reversed-Phase High-Performance Liquid Chromatography in Cultured Cells. Calcium Signal. 2019, 1925, 223–232. [Google Scholar] [CrossRef]
- Barraud, C.; Corbière, C.; Pottier, I.; Estace, E.; Blanchard, K.; Logie, C.; Lagadu, S.; Kéravec, V.; Dionnet, F.; Morin, J.; et al. Impact of after-treatment devices and biofuels on diesel exhausts genotoxicity in A549 cells exposed at air-liquid interface. Toxicol. In Vitro 2017, 45, 426–433. [Google Scholar] [CrossRef] [PubMed]
- Qiu, W.-Q.; Chen, S.-S.; Xie, J.; Qu, Y.-H.; Song, X. Analysis of 10 nucleotides and related compounds in Litopenaeus vannamei during chilled storage by HPLC-DAD. LWT 2016, 67, 187–193. [Google Scholar] [CrossRef]
- Nguyen, L.H.; Chong, N.-M. Development of an ATP measurement method suitable for xenobiotic treatment activated sludge biomass. J. Chromatogr. B 2015, 1000, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Akhova, A.V.; Tkachenko, A.G. HPLC–UV method for simultaneous determination of adenosine triphosphate and its metabolites inMycobacterium smegmatis. Acta Chromatogr. 2019, 31, 45–48. [Google Scholar] [CrossRef]
- García-Tardón, N.; Guigas, B. Determination of Adenine Nucleotide Concentrations in Cells and Tissues by High-Performance Liquid Chromatography. Methods Mol. Biol. 2018, 1732, 229–237. [Google Scholar] [PubMed]
- Feng, J.H.; Wei, K.Z.; Gao, J.P.; Xu, X. Determination of adenosine phosphates in mouse myocardium tissue by HPLC with UV detection and using porous graphite carbon column. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2020, 1145, 122110. [Google Scholar] [CrossRef] [PubMed]
- Juarez-Facio, A.T.; Castilla, C.; Corbière, C.; Lavanant, H.; Afonso, C.; Morin, C.; Merlet-Machour, N.; Chevalier, L.; Vaugeois, J.-M.; Yon, J.; et al. Development of a standardized in vitro approach to evaluate microphysical, chemical, and toxicological properties of combustion-derived fine and ultrafine particles. J. Environ. Sci. 2021, 113, 104–117. [Google Scholar] [CrossRef]
- ICH Harmonized Tripartite Guideline: Validation of Analytical Procedures: Text and Methodology Q2(R1). In Proceedings of the International Conference of Harmonization of Technical Requirements for Registration of Pharmaceuticals for Human Use, Geneva, Switzerland, November 2005; Available online: https://www.ema.europea.eu/en/documents/scientific-guideline/ich-q-2-r1-validation-analytical-procedures-text-methodology-step-5_en.pdf (accessed on 19 September 2021).


| Analytes | Calibration Curve | Correlation Coefficient R2 | Linear Ranger (µM) | LOD (µM) | LOQ (µM) | Recovery (%) | CV% |
|---|---|---|---|---|---|---|---|
| ATP | y = 35.21x − 0.7885 | 0.9999 | 0.2–10 | 0.054 | 0.18 | 110.4 | 2.9 |
| ADP | y = 28.9x − 0.1143 | 0.9999 | 0.2–10 | 0.060 | 0.20 | 97.8 | 8.1 |
| AMP | y = 30.5x − 0.5111 | 0.9999 | 0.2–10 | 0.051 | 0.17 | 110.5 | 5.1 |
| Analytes | Concentration (µM) | Intra-Day (n = 3) | Inter-Day (n = 9) | ||||
|---|---|---|---|---|---|---|---|
| Mean Measured Concentration (µM) ± S.D. | Bias% | CV% | Mean Measured Concentration (µM) ± S.D. | Bias% | CV% | ||
| ATP | 0.2 | 0.22 ± 0.001 | 11.3 | 0.7 | 0.20 ± 0.02 | 0.1 | 8.7 |
| 0.6 | 0.63 ± 0.003 | 4.3 | 0.5 | 0.61 ± 0.01 | 2.2 | 2.1 | |
| 5 | 5.00 ± 0.016 | 0.8 | 0.4 | 5.03 ± 0.03 | 0.7 | 0.5 | |
| 7.5 | 7.42 ± 0.194 | 1.1 | 2.6 | 7.50 ± 0.13 | 0.1 | 1.7 | |
| ADP | 0.2 | 0.20 ± 0.006 | 1.9 | 3.1 | 0.19 ± 0.01 | 3.7 | 4.8 |
| 0.6 | 0.60 ± 0.003 | 0.2 | 0.5 | 0.60 ± 0.01 | 0.3 | 1.1 | |
| 5 | 4.95 ± 0.015 | 1.1 | 0.3 | 4.97 ± 0.02 | 0.5 | 0.5 | |
| 7.5 | 7.29 ± 0.193 | 2.7 | 2.6 | 7.44 ± 0.15 | 0.9 | 1.7 | |
| AMP | 0.2 | 0.22 ± 0.001 | 8.1 | 0.2 | 0.20 ± 0.01 | 0.2 | 6.5 |
| 0.6 | 0.57 ± 0.020 | 2.5 | 0.6 | 0.61 ± 0.01 | 2.4 | 0.5 | |
| 5 | 5.00 ± 0.020 | 0.1 | 0.4 | 5.06 ± 0.06 | 1.1 | 1.2 | |
| 7.5 | 7.37 ± 0.195 | 1.8 | 2.7 | 7.52 ± 0.16 | 0.3 | 2.2 | |
| Sample No. | ATP | ADP | AMP | ATP/ADP |
|---|---|---|---|---|
| 1 | 3.90 × 10−5 | 1.43 × 10−6 | 7.91 × 10−7 | 27.3 |
| 2 | 3.43 × 10−5 | 1.26 × 10−6 | 1.47 × 10−7 | 27.2 |
| 3 | 3.40 × 10−5 | 1.22 × 10−6 | 1.76 × 10−6 | 27.9 |
| 4 | 3.52 × 10−5 | 1.26 × 10−6 | 7.04 × 10−7 | 27.9 |
| Mean | 3.56 × 10−5 | 1.29 × 10−6 | 8.51 × 10−7 | 27.6 |
| SD | 2.31 × 10−6 | 9.36 × 10−8 | 6.70 × 10−7 | 0.4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Juarez-Facio, A.T.; Martin de Lagarde, V.; Monteil, C.; Vaugeois, J.-M.; Corbiere, C.; Rogez-Florent, T. Validation of a Fast and Simple HPLC-UV Method for the Quantification of Adenosine Phosphates in Human Bronchial Epithelial Cells. Molecules 2021, 26, 6324. https://doi.org/10.3390/molecules26206324
Juarez-Facio AT, Martin de Lagarde V, Monteil C, Vaugeois J-M, Corbiere C, Rogez-Florent T. Validation of a Fast and Simple HPLC-UV Method for the Quantification of Adenosine Phosphates in Human Bronchial Epithelial Cells. Molecules. 2021; 26(20):6324. https://doi.org/10.3390/molecules26206324
Chicago/Turabian StyleJuarez-Facio, Ana Teresa, Violaine Martin de Lagarde, Christelle Monteil, Jean-Marie Vaugeois, Cécile Corbiere, and Tiphaine Rogez-Florent. 2021. "Validation of a Fast and Simple HPLC-UV Method for the Quantification of Adenosine Phosphates in Human Bronchial Epithelial Cells" Molecules 26, no. 20: 6324. https://doi.org/10.3390/molecules26206324
APA StyleJuarez-Facio, A. T., Martin de Lagarde, V., Monteil, C., Vaugeois, J. -M., Corbiere, C., & Rogez-Florent, T. (2021). Validation of a Fast and Simple HPLC-UV Method for the Quantification of Adenosine Phosphates in Human Bronchial Epithelial Cells. Molecules, 26(20), 6324. https://doi.org/10.3390/molecules26206324