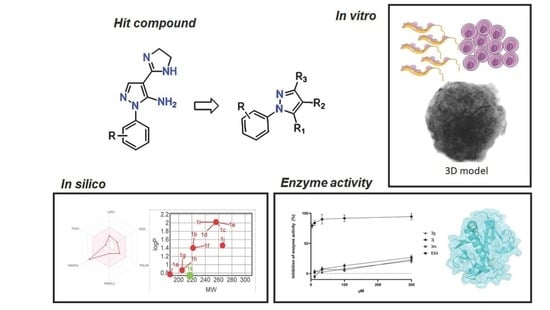
Structural Optimization and Biological Activity of Pyrazole Derivatives: Virtual Computational Analysis, Recovery Assay and 3D Culture Model as Potential Predictive Tools of Effectiveness against Trypanosoma cruzi
Abstract
:1. Introduction
2. Results and Discussion
2.1. Drug Design and Synthesis
2.2. Physicochemical Properties Prediction
2.3. Toxicity and Trypanocidal Effect of Pyrazole Derivatives
2.4. Structure-Activity Relationship (SAR) of Pyrazole Derivatives
2.5. Effect of Pyrazole Derivatives on Parasite Recrudescence
2.6. 3D Culture Model as Potential Drug Efficacy Prediction
2.7. ADMET Analysis
2.8. Enzyme Activity
3. Materials and Methods
3.1. Compound Synthesis
- 1-aryl-1H-pyrazole-carboxamides 1(a-l) and 5-amino-1-aryl-1H-pyrazole-carboxamides 2(a–l)
- 1-phenyl-1H-pyrazole-4-carboxamide (1a)
- 1-(3-chlorophenyl)-1H-pyrazole-4-carboxamide (1b)
- 1-(2,4-dichlorophenyl)-1H-pyrazole-4-carboxamide (1c)
- 1-(3,5-dichlorophenyl)-1H-pyrazole-4-carboxamide (1d)
- 1-(3,4-dichlorophenyl)-1H-pyrazole-4-carboxamide (1e)
- 1-(4-chlorophenyl)-1H-pyrazole-4-carboxamide (1f)
- 1-(4-fluorophenyl)-1H-pyrazole-4-carboxamide (1g)
- 1-(3-fluorophenyl)-1H-pyrazole-4-carboxamide (1h)
- 1-(4-bromophenyl)-1H-pyrazole-4-carboxamide (1i)
- 1-(3-bromophenyl)-1H-pyrazole-4-carboxamide (1j)
- 1-(4-methoxyphenyl)-1H-pyrazole-4-carboxamide (1k)
- 1-(2,3-dichlorophenyl)-1H-pyrazole-4-carboxamide (1l)
- 5-amino-1-phenyl-1H-pyrazole-4-carboxamide (2a)
- 5-amino-1-(3-chlorophenyl)-1H-pyrazole-4-carboxamide (2b)
- 5-amino-1-(2,4-dichlorophenyl)-1H-pyrazole-4-carboxamide (2c)
- 5-amino-1-(3,5-dichlorophenyl)-1H-pyrazole-4-carboxamide (2d)
- 5-amino-1-(3,4-dichlorophenyl)-1H-pyrazole-4-carboxamide (2e)
- 5-amino-1-(4-chlorophenyl)-1H-pyrazole-4-carboxamide (2f)
- 5-amino-1-(4-fluorophenyl)-1H-pyrazole-4-carboxamide (2g)
- 5-amino-1-(3-fluorophenyl)-1H-pyrazole-4-carboxamide (2h)
- 5-amino-1-(4-bromophenyl)-1H-pyrazole-4-carboxamide (2i)
- 5-amino-1-(3-bromophenyl)-1H-pyrazole-4-carboxamide (2j)
- 5-amino-1-(4-methoxyphenyl)-1H-pyrazole-4-carboxamide (2k)
- 5-amino-1-(2,3-dichlorophenyl)-1H-pyrazole-4-carboxamide (2l)
- 1-aryl-4-(4,5-dihydro-1H-imidazol-2-yl)-1H-pyrazoles3(a–n)
- 4-(4,5-dihydro-1H-imidazol-2-yl)-1-phenyl-1H-pyrazole (3a)
- 1-(3-chlorophenyl)-4-(4,5-dihydro-1H-imidazol-2-yl)-1H-pyrazole (3b)
- 1-(2,4-dichlorophenyl)-4-(4,5-dihydro-1H-imidazol-2-yl)-1H-pyrazole (3c)
- 1-(3,5-dichlorophenyl)-4-(4,5-dihydro-1H-imidazol-2-yl)-1H-pyrazole (3d)
- 1-(3,4-dichlorophenyl)-4-(4,5-dihydro-1H-imidazol-2-yl)-1H-pyrazole (3e)
- 1-(2,6-dichlorophenyl)-4-(4,5-dihydro-1H-imidazol-2-yl)-1H-pyrazole (3f)
- 1-(4-chlorophenyl)-4-(4,5-dihydro-1H-imidazol-2-yl)-1H-pyrazole (3g)
- 4-(4,5-dihydro-1H-imidazol-2-yl)-1-(4-fluorophenyl)-1H-pyrazole (3h)
- 4-(4,5-dihydro-1H-imidazol-2-yl)-1-(3-fluorophenyl)-1H-pyrazole (3i)
- 1-(4-bromophenyl)-4-(4,5-dihydro-1H-imidazol-2-yl)-1H-pyrazole (3j)
- 1-(3-bromophenyl)-4-(4,5-dihydro-1H-imidazol-2-yl)-1H-pyrazole (3k)
- 4-(4,5-dihydro-1H-imidazol-2-yl)-1-(4-methoxyphenyl)-1H-pyrazole (3l)
- 1-(3-chloro-4-methylphenyl)-4-(4,5-dihydro-1H-imidazol-2-yl)-1H-pyrazole (3m)
- 1-(4-chloro-2-methylphenyl)-4-(4,5-dihydro-1H-imidazol-2-yl)-1H-pyrazole (3n)
- 5-amino-1-aryl-3-methyl-4-(4,5-dihydro-1H-imidazol-2-yl)-1H-pyrazoles4(a–c)
- 5-amino-1-(3-chlorophenyl)-3-methyl-4-(4,5-dihydro-1H-imidazol-2-yl)-1H-pyrazole (4a)
- 5-amino-1-(3,5-dichlorophenyl)-3-methyl-4-(4,5-dihydro-1H-imidazol-2-yl)-1H-pyrazole (4b)
- 5-amino-1-(3,4-dichlorophenyl)-3-methyl-4-(4,5-dihydro-1H-imidazol-2-yl)-1H-pyrazole (4c)
- 5-amino-1-aryl-4-(4(5)-methyl-4,5-dihydro-1H-imidazol-2-yl)-1H-pyrazoles5(a–c)
- 5-amino-1-(3-chlorophenyl)-4-(4-(5)-methyl-4,5-dihydro-1H-imidazol-2-yl)-1H-pyrazole (5a)
- 5-amino-1-(3,5-dichlorophenyl)-4-(4-(5)-methyl-4,5-dihydro-1H-imidazol-2-yl)-1H-pyrazole (5b)
- 5-amino-1-(3,4-dichlorophenyl)-4-(-4(5)-methyl-4,5-dihydro-1H-imidazol-2-yl)-1H-pyrazole (5c)
3.2. Cell Culture
3.2.1. Two-Dimensional Culture (2D)
3.2.2. Three-Dimensional Culture (3D)
3.3. Parasites and Culture Infection
3.4. Cytotoxicity In Vitro Assay
3.5. Anti-T. cruzi Compound Screening
3.6. Fluorescence Microscopy
3.7. Reversibility Assay (Washout)
3.8. Enzyme Activity in Solution
3.9. Physicochemical and ADMET Prediction
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- World Health Organization. Neglected Tropical Disease. 2020. Available online: https://www.who.int/neglected_diseases/diseases/en/ (accessed on 21 September 2021).
- Antinori, S.; Galimberti, L.; Bianco, R.; Grande, R.; Gali, M.; Corbellino, M. Chagas disease in Europe: A review for the internist in the globalized world. Eur. J. Intern. Med. 2017, 43, 6–15. [Google Scholar] [CrossRef]
- World Health Organization. Chagas Disease. 2020. Available online: https://www.who.int/news-room/fact-sheets/detail/chagas-disease-(american-trypanosomiasis) (accessed on 21 September 2021).
- Rassi, A., Jr.; Rassi, A.; Marin-Neto, J.A. Chagas disease. Lancet 2010, 375, 1388–1402. [Google Scholar] [CrossRef]
- Rassi, A., Jr.; Rassi, A.; Marcondes de Rezende, J. American trypanosomiasis (Chagas disease). Infect. Dis. Clin. N. Am. 2012, 26, 275–291. [Google Scholar] [CrossRef]
- Lidani, K.C.F.; Andrade, F.A.; Bavia, L.; Damasceno, F.S.; Beltrame, M.H.; Messias-Reason, I.J.; Sandri, T.L. Chagas disease: From discovery to a worldwide health problem. Front. Public Health 2019, 7, 166–179. [Google Scholar] [CrossRef] [PubMed]
- Petravicius, P.O.; Costa-Martins, A.G.; Silva, M.N.; Reis-Cunha, J.L.; Bartholomeu, D.C.; Teixeira, M.M.G.; Zingales, B. Mapping benznidazole resistance in trypanosomatids and exploring evolutionary histories of nitroreductases and ABCG transporter protein sequences. Acta Trop. 2019, 200, 105161–105171. [Google Scholar] [CrossRef]
- Morillo, C.A.; Marin-Neto, J.A.; Avezum, A.; Sosa-Estani, S.; Rassi, A., Jr.; Rosas, F.; Villena, E.; Quiroz, R.; Bonilla, R.; Britto, C.; et al. Randomized trial of benznidazole for chronic Chagas’ cardiomyopathy. N. Engl. J. Med. 2015, 373, 1295–1306. [Google Scholar] [CrossRef] [Green Version]
- Molina, I.; Prat, J.G.; Salvador, F.; Treviño, B.; Sulleiro, E.; Serre, N.; Pou, D.; Roure, S.; Cabezos, J.; Valerio, L.; et al. Randomized trial of posaconazole and benznidazole for chronic Chagas’ disease. N. Engl. J. Med. 2014, 370, 1899–1908. [Google Scholar] [CrossRef] [PubMed]
- Morillo, C.A.; Waskin, H.; Sosa-Estani, S.; Del Carmen Bangher, M.; Cuneo, C.; Milesi, R.; Mallagray, M.; Apt, W.; Beloscar, J.; Gascon, J.; et al. Benznidazole and posaconazole in eliminating parasites in asymptomatic T. cruzi carriers. J. Am. Coll. Cardiol. 2017, 69, 939–947. [Google Scholar] [CrossRef]
- Torrico, F.; Gascon, J.; Oritz, L.; Alonso-Veja, C.; Pinazo, M.J.; Schijman, A.; Almeida, A.C.; Alves, F.; Strub-Wourgaft, N.; Ribeiro, I.; et al. Treatment of adult chronic indeterminate Chagas disease with benznidazole and three E1224 dosing regimens: A proof-of-concept, randomised, placebo-controlled trial. Lancet Infect. Dis. 2018, 18, 419–430. [Google Scholar] [CrossRef]
- Torrico, F.; Gascón, J.; Barreira, F.; Blum, B.; Almeida, I.C.; Alonso-Vega, C.; Barboza, T.; Bilbe, G.; Correia, E.; Garcia, W.; et al. New regimens of benznidazole monotherapy and in combination with fosravuconazole for treatment of Chagas disease (BENDITA): A phase 2, double-blind, randomised trial. Lancet Infect. Dis. 2021, 21, 1129–1140. [Google Scholar] [CrossRef]
- Monteiro, M.E.; Lechuga, G.; Lara, L.S.; Souto, B.A.; Viganó, M.G.; Bourguignon, S.C.; Calvet, C.M.; Oliveira, F.O.R., Jr.; Alves, C.R.; Souza-Silva, F.; et al. Synthesis, structure-activity relationship and trypanocidal activity of pyrazole-imidazoline and new pyrazole-tetrahydropyrimidine hybrids as promising chemotherapeutic agents for Chagas disease. Eur. J. Med. Chem. 2019, 182, 111610–111623. [Google Scholar] [CrossRef]
- Scharfstein, J.; Schmitz, V.; Morandi, V.; Capella, M.M.; Lima, A.P.; Morrot, A.; Juliano, L.; Muller-Esterl, W. Host cell invasion by Trypanosoma cruzi is potentiated by activation of bradykinin B(2) receptors. J. Exp. Med. 2000, 192, 1289–1300. [Google Scholar] [CrossRef]
- Meirelles, M.N.; Juliano, L.; Carmona, E.; Silva, S.G.; Costa, E.M.; Murta, A.C.; Scharfstein, J. Inhibitors of the major cysteinyl proteinase (GP57/51) impair host cell invasion and arrest the intracellular development of Trypanosoma cruzi in vitro. Mol. Biochem. Parasitol. 1992, 52, 175–184. [Google Scholar] [CrossRef]
- Santos, C.D.; Caldeira, J.C.; Toldo, M.P.A.; Prado, J.C. Trypanosoma cruzi: Effects of repetitive stress during the development of experimental infection. Exp. Parasitol. 2005, 110, 96–101. [Google Scholar] [CrossRef]
- Doyle, P.S.; Zhou, Y.M.; Hsieh, I.; Greenbaum, D.C.; McKerrow, J.H.; Engel, J.C. The Trypanosoma cruzi protease cruzain mediates immune evasion. PLoS Pathog. 2011, 7, e1002139. [Google Scholar] [CrossRef] [Green Version]
- DNDi. K777 (Chagas). 2014. Available online: https://dndi.org/research-development/portfolio/k777/ (accessed on 21 September 2021).
- McKerrow, J.H. Update on drug development targeting parasite cysteine proteases. PLoS Negl. Trop. Dis. 2018, 12, e0005850. [Google Scholar] [CrossRef]
- Beaulieu, C.; Isabel, E.; Fortier, A.; Massé, F.; Mellon, C.; Méthot, N.; Ndao, M.; Nicoll-Griffith, D.; Lee, D.; Park, H.; et al. Identification of potent and reversible cruzipain inhibitors for the treatment of Chagas disease. Bioorg. Med. Chem. Lett. 2010, 20, 7444–7449. [Google Scholar] [CrossRef] [PubMed]
- Ndao, M.; Beaulieu, C.; Black, W.C.; Isabel, E.; Vasquez-Camargo, F.; Nath-Chowdhury, M.; Massé, F.; Mellon, C.; Methot, N.; Nicoll-Griffith, D.A. Reversible cysteine protease inhibitors show promise for a Chagas disease cure. Antimicrob. Agents Chemother. 2014, 58, 1167–1178. [Google Scholar] [CrossRef] [Green Version]
- Salas-Sarduy, E.; Landaburu, L.U.; Karpiak, J.; Madauss, K.P.; Cazzulo, J.J.; Aguero, F.; Alvarez, V.E. Novel scaffolds for inhibition of cruzipain identified from high-throughput screening of anti-kinetoplastid chemical boxes. Sci. Rep. 2017, 7, 12073–12085. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palos, I.; Lara-Ramirez, E.E.; Lopez-Cedillo, J.C.; Garcia-Perez, C.; Kashif, M.; Bocanegra-Garcia, V.; Nogueda-Torres, B.; Rivera, G. Repositioning FDA drugs as potential cruzain inhibitors from Trypanosoma cruzi: Virtual screening, in vitro and in vivo studies. Molecules 2017, 22, 1015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tochowicz, A.; Mckerrow, J.H.; Craik, C.S. Crystal Structure Analysis of Cruzain with Fragment 1 (N-(1H-benimidazole-2-yl)-1,3-dimethyl-pyrazole-4-carboxamide). Available online: https://www.wwpdb.org/pdb?id=pdb_00004w5b (accessed on 21 September 2021).
- Leung, C.S.; Leung, S.S.F.; Tirado-Rives, J.; Jorgensen, W.L. Methyl effects on protein-ligand binding. J. Med. Chem. 2012, 55, 4489–4500. [Google Scholar] [CrossRef] [Green Version]
- Ferreira, B.S.; Silva, R.C.; Souto, B.A.; Santos, M.S. Synthesis of pyrazole-carboxamides and pyrazole-carboxylic acids derivatives: Simple methods to access powerful building blocks. Lett. Org. Chem. 2021, 18, 335–343. [Google Scholar] [CrossRef]
- Rosa, G.S.; Souto, B.A.; Pereira, C.N.; Teixeira, B.C.; Santos, M.S. A convenient synthesis of pyrazole-imidazoline derivatives by microwave irradiation. J. Heterocycl. Chem. 2019, 56, 1825–1830. [Google Scholar] [CrossRef]
- Santos, M.S.; Bernardino, A.M.R.; Pinheiro, L.C.S.; Canto-Cavalheiro, M.M.; Leon, L.L. An efficient synthesis of new 5-(1-Aryl-1H-pyrazole-4-yl)-1H-tetrazoles from 1-Aryl-1H-pyrazole-4-carbonitriles via [3 + 2] cycloaddition reaction. J. Heterocycl. Chem. 2012, 49, 1425–1428. [Google Scholar] [CrossRef]
- Faria, J.V.; Santos, M.S.; Bernardino, A.M.R.; Becker, K.M.; Machado, G.M.C.; Rodrigues, R.F.; Canto-Cavalheiro, M.M.; Leon, L.L. Synthesis and activity of novel tetrazole compounds and their pyrazole-4-carbonitrile precursors against Leishmania spp. Bioorg. Med. Chem. Lett. 2013, 23, 6310–6312. [Google Scholar] [CrossRef]
- Santos, M.S.; Oliveira, M.L.; Bernardino, A.M.; de Léo, R.M.; Amaral, V.F.; de Carvalho, F.T.; Leon, L.L.; Canto-Cavalheiro, M.M. Synthesis and antileishmanial evaluation of 1-aryl-4-(4,5-dihydro-1H-imidazol-2-yl)-1H-pyrazole derivatives. Bioorg. Med. Chem. Lett. 2011, 21, 7451–7454. [Google Scholar] [CrossRef]
- Bunally, S.B.; Luscombe, C.N.; Young, R.J. Using physicochemical measurements to influence better compound design. SLAS Discov. 2019, 24, 791–801. [Google Scholar] [CrossRef]
- Chandrasekaran, B.; Abed, S.N.; Al-Attraqchi, O.; Kuche, K.; Tekade, R.K. Computer-aided prediction of pharmacokinetic (ADMET) properties. In Advances in Pharmaceutical Product Development and Research: Dosage form Design Parameters; Tekade, R.K., Ed.; Academic Press: Cambridge, MA, USA, 2018; Volume 2, pp. 731–755. [Google Scholar]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717–42730. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peters, J.U.; Schnider, P.; Mattei, P.; Kansy, M. Pharmacological promiscuity: Dependence on compound properties and target specificity in a set of recent roche compounds. Chem. Med. Chem. 2009, 4, 680–686. [Google Scholar] [CrossRef] [PubMed]
- Salvador, R.R.S.; Bello, M.L.; Barreto, I.R.L.; Vera, M.A.F.; Muri, E.M.F.; Albuquerque, S.D.E.; Dias, L.R.S. New carbohydrazide derivatives of 1H-pyrazolo[3,4-b]pyridine and trypanocidal activity. An. Acad. Bras. Cienc. 2016, 88, 2341–2348. [Google Scholar] [CrossRef] [Green Version]
- MacLean, L.M.; Thomas, J.; Lewis, M.D.; Cotillo, I.; Gray, D.W.; Rycker, M. Development of Trypanosoma cruzi in vitro assays to identify compounds suitable for progression in Chagas’ disease drug Discovery. PLoS Negl. Trop. Dis. 2018, 12, e0006612. [Google Scholar] [CrossRef]
- Varghese, S.; Rahmani, R.; Russel, S.; Deora, G.S.; Ferrins, L.; Toynton, A.; Jones, A.; Sykes, M.; Kessler, A.; Eufrásio, A.; et al. Discovery of potent N-ethylurea pyrazole derivatives as dual inhibitors of Trypanosoma brucei and Trypanosoma cruzi. ACS Med. Chem. Lett. 2019, 11, 278–285. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; Hulpia, F.; Silva, C.F.; Batista, D.G.J.; Hecke, K.V.; Maes, L.; Caljon, G.; Soeiro, M.N.C.; Calenbergh, S.V. Discovery of pyrrolo[2,3-b]pyridine (1,7-Dideazapurine) Nucleoside analogues as anti-Trypanosoma cruzi agents. J. Med. Chem. 2019, 62, 8847–8865. [Google Scholar] [CrossRef]
- Sykes, M.L.; Avery, V.M. 3-pyridyl inhibitors with novel activity against Trypanosoma cruzi reveal in vitro profiles can aid prediction of putative cytochrome P450 inhibition. Sci. Rep. 2018, 8, 4901–4913. [Google Scholar] [CrossRef] [Green Version]
- Langhans, S.A. Three-Dimensional in vitro cell culture models in drug discovery and drug repositioning. Front. Pharmacol. 2019, 9, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, M.E.; Rizzi, M.; Caeiro, L.D.; Masip, Y.E.; Perrone, A.; Sánchez, D.O.; Búa, J.; Tekiel, V. Transmigration of Trypanosoma cruzi trypomastigotes through 3D cultures resembling a physiological environment. Cell. Microbiol. 2020, 22, e13207. [Google Scholar] [CrossRef]
- Garzoni, L.R.; Adesse, D.; Soares, M.J.; Rossi, M.I.D.; Borojevic, R.; Meirelles, M.N.L. Fibrosis and hypertrophy induced by Trypanosoma cruzi in a three-dimensional cardiomyocyte-culture system. J. Infect. Dis. 2008, 197, 906–915. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nisimura, L.M.; Ferrão, P.M.; Nogueira, A.R.; Waghabi, M.C.; Meuser-Batista, M.; Moreira, O.C.; Urbina, J.A.; Garzoni, L.R. Effect of posaconazole in an in vitro model of cardiac fibrosis induced by Trypanosoma cruzi. Mol. Biochem. Parasitol. 2020, 238, 111283. [Google Scholar] [CrossRef]
- Arez, F.; Rebelo, S.P.; Fontinha, D.; Simão, D.; Martins, T.R.; Machado, M.; Fischli, C.; Oeuvray, C.; Badolo, L.; Carrondo, M.J.T.; et al. Flexible 3D cell-based platforms for the discovery and profiling of novel drugs targeting Plasmodium hepatic infection. ACS Infect. Dis. 2019, 5, 1831–1842. [Google Scholar] [CrossRef]
- Koch, J.; Monch, D.; Maa, A.; Gromoll, C.; Hehr, T.; Leibold, T.; Schlitt, H.J.; Dahlke, M.; Renner, P. Three-dimensional cultivation increases chemo- and radioresistance of colorectal cancer cell lines. PLoS ONE 2021, 16, e0244513. [Google Scholar] [CrossRef]
- Reviriego, F.; Olmo, F.; Navarro, P.; Marín, C.; Ramírez-Macías, I.; García-España, E.; Albelda, M.T.; Gutiérrez-Sánchez, R.; Sánchez-Moreno, M.; Arán, V.J. Simple dialkyl pyrazole-3,5-dicarboxylates show in vitro and in vivo activity against disease-causing trypanosomatids. Parasitology 2017, 144, 1133–1143. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Moreno, M.; Marín, C.; Navarro, P.; Lamarque, L.; García-España, E.; Miranda, C.; Huertas, O.; Olmo, F.; Gómez-Contreras, F.; Pitarch, J.; et al. In vitro and in vivo trypanosomicidal activity of pyrazole-containing macrocyclic and macrobicyclic polyamines: Their action on acute and chronic phases of Chagas disease. J. Med. Chem. 2012, 55, 4231–4243. [Google Scholar] [CrossRef]
- Fiuza, L.F.A.; Peres, R.B.; Simões-Silva, M.R.; Silva, P.B.; Batista, D.G.J.; Silva, C.F.; Gama, A.N.S.; Reddy, T.R.K.; Soeiro, M.N.C. Identification of Pyrazolo[3,4-e][1,4]thiazepin based CYP51 inhibitors as potential Chagas disease therapeutic alternative: In vitro and in vivo evaluation, binding mode prediction and SAR exploration. Eur. J. Med. Chem. 2018, 149, 257–268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ammerman, N.C.; Beier-Sexton, M.; Azad, A.F. Growth and Maintenance of Vero Cell Lines. Curr. Protoc. Microbiol. 2008, 11, A.4E.1–A.4E.7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henriques, C.; Castro, D.P.; Gomes, L.H.F.; Garcia, E.S.; de Souza, W. Bioluminescent imaging of Trypanosoma cruzi infection in Rhodnius prolixus. Parasites Vectors 2012, 5, 214–229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sander, T.; Freyss, J.; von Korff, M.; DataWarrior, R.C. An open-source program for chemistry aware data visualization and analysis. J. Chem. Inf. Model. 2015, 55, 460–473. [Google Scholar] [CrossRef] [PubMed]
- Lagorce, D.; Sperandio, O.; Baell, J.B.; Miteva, M.A.; Villoutreix, B.O. FAF-Drugs3: A web server for compound property calculation and chemical library design. Nucleic Acids Res. 2015, 43, W200–W207. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Lou, C.; Sun, L.; Li, J.; Cai, Y.; Wang, Z.; Li, W.; Liu, G.; Tang, Y. admetSAR 2.0: Web-service for prediction and optimization of chemical ADMET properties. Bioinformatics 2019, 35, 1067–1069. [Google Scholar] [CrossRef] [PubMed]







| Compounds | Trypanocidal Activity (Mean ± SD μM) | Cytotoxicity (Mean ± SD μM) | |||||
|---|---|---|---|---|---|---|---|
| Trypomastigotes | Intracellular Amastigotes | ||||||
| IC50 | IC90 | SI | IC50 | IC90 | SI | CC50 | |
| Series 1(a–l) | >100 | Nd | Nd | >50 | Nd | Nd | >500 |
| Series 2(a–l) | >100 | Nd | Nd | >70 | Nd | Nd | >500 |
| 3a | >100 | Nd | Nd | 91.82 ± 2.03 | >100 | >5.44 | >500 |
| 3b | >100 | Nd | Nd | 28.16 ± 1.81 | 86.60 ± 5.55 | 9.48 | 267.10 ± 28.15 |
| 3c | >100 | Nd | Nd | 56.64 ± 2.52 | 94.61 ± 3.61 | 7.21 | 408.69 ± 16.17 |
| 3d | 66.30 ± 5.53 | 93.48 ± 1.56 | 5.78 | 54.91 ± 7.12 | >100 | 6.99 | 383.83 ± 22.97 |
| 3e | 64.86 ± 2.85 | 93.74 ± 0.73 | 7.39 | 76.52 ± 8.79 | >100 | 6.26 | 479.66 ± 21.67 |
| 3f | >100 | Nd | Nd | 82.00 ± 2.52 | >100 | >6 | >500 |
| 3g | 64.60 ± 1.56 | >100 | 4.27 | 6.09 ± 0.52 | 26.10 ± 14.31 | 45.52 | 277.24 ± 15.76 |
| 3h | >100 | Nd | Nd | 64.50 ± 3.81 | >100 | 7.80 | >500 |
| 3i | >100 | Nd | Nd | 64.12 ± 7.09 | >100 | 7.80 | >500 |
| 3j | 57.71 ± 3.14 | 95.32 ± 5.43 | 5.36 | 2.75 ± 0.62 | 9.67 ± 0.30 | 112.48 | 309.33 ± 34.31 |
| 3k | >100 | Nd | Nd | 26.92 ± 2.19 | 72.85 ± 3.67 | 7.84 | 211.04 ± 31.68 |
| 3l | >100 | Nd | Nd | 71.76 ± 2.71 | >100 | >7 | >500 |
| 3m | 34.54 ± 8.32 | >100 | 4.64 | 3.58 ± 0.25 | 21.37 ± 1.25 | 44.83 | 160.51 ± 16.13 |
| 3n | >100 | Nd | Nd | 24.68 ± 3.42 | 85.72 ± 5.03 | 13.86 | 342.17 ± 29.99 |
| 4a | >100 | >100 | Nd | >100 | >100 | >5 | >500 |
| 4b | >100 | >100 | Nd | 90.67 ± 13.17 | >100 | >5.51 | >500 |
| 4c | >100 | >100 | Nd | 93.83 ± 16.40 | >100 | 3.78 | 355.4 ± 17.72 |
| 5a | >100 | >100 | Nd | >100 | >100 | Nd | >500 |
| 5b | >100 | >100 | Nd | 32.95 ± 8.84 | >100 | >15.17 | >500 |
| 5c | >100 | >100 | Nd | 34.33 ± 4.24 | >100 | >9.98 | 342.93 ± 37.0 |
| Bz | 18.71 ± 4.58 | >100 | >26.7 | 4.67 ± 0.22 | >100 | >107 | >500 |
 | ||||||
|---|---|---|---|---|---|---|
| Compounds | Structures | R1 | R2 | R3 | R4 | Intracellular Amastigotes |
| pIC50 | ||||||
| 1a |  | H | H | C(O)NH2 | H | <4.3 |
| 1b |  | H | 3-Cl | C(O)NH2 | H | <4.3 |
| 1c |  | H | 2,4-diCl | C(O)NH2 | H | <4.3 |
| 1d |  | H | 3,5-diCl | C(O)NH2 | H | <4.3 |
| 1e |  | H | 3,4-diCl | C(O)NH2 | H | <4.3 |
| 1f |  | H | 4-Cl | C(O)NH2 | H | <4.3 |
| 1g |  | H | 4-F | C(O)NH2 | H | <4.3 |
| 1h |  | H | 3-F | C(O)NH2 | H | <4.3 |
| 1i |  | H | 4-Br | C(O)NH2 | H | <4.3 |
| 1j |  | H | 3-Br | C(O)NH2 | H | <4.3 |
| 1k |  | H | 4-OCH3 | C(O)NH2 | H | <4.3 |
| 1l |  | H | 2,3-diCl | C(O)NH2 | H | <4.3 |
| 2a |  | NH2 | H | C(O)NH2 | H | <4.15 |
| 2b |  | NH2 | 3-Cl | C(O)NH2 | H | <4.15 |
| 2c |  | NH2 | 2,4-diCl | C(O)NH2 | H | <4.15 |
| 2d |  | NH2 | 3,5-diCl | C(O)NH2 | H | <4.15 |
| 2e |  | NH2 | 3,4-diCl | C(O)NH2 | H | <4.15 |
| 2f |  | NH2 | 4-Cl | C(O)NH2 | H | <4.15 |
| 2g |  | NH2 | 4-F | C(O)NH2 | H | <4.15 |
| 2h |  | NH2 | 3-F | C(O)NH2 | H | <4.15 |
| 2i |  | NH2 | 4-Br | C(O)NH2 | H | <4.15 |
| 2j |  | NH2 | 3-Br | C(O)NH2 | H | <4.15 |
| 2k |  | NH2 | 4-OCH3 | C(O)NH2 | H | <4.15 |
| 2l |  | NH2 | 2,3-diCl | C(O)NH2 | H | <4.15 |
| 3a |  | H | H | C3H5N2 | H | 4.03 |
| 3b |  | H | 3-Cl | C3H5N2 | H | 4.55 |
| 3c |  | H | 2,4-diCl | C3H5N2 | H | 4.25 |
| 3d |  | H | 3,5-diCl | C3H5N2 | H | 4.26 |
| 3e |  | H | 3,4-diCl | C3H5N2 | H | 4.12 |
| 3f |  | H | 2,6-diCl | C3H5N2 | H | 4.09 |
| 3g |  | H | 4-Cl | C3H5N2 | H | 5.22 |
| 3h |  | H | 4-F | C3H5N2 | H | 4.19 |
| 3i |  | H | 3-F | C3H5N2 | H | 4.19 |
| 3j |  | H | 4-Br | C3H5N2 | H | 5.56 |
| 3k |  | H | 3-Br | C3H5N2 | H | 4.57 |
| 3l |  | H | 4-OCH3 | C3H5N2 | H | 4.14 |
| 3m |  | H | 3-Cl,4-CH3 | C3H5N2 | H | 5.45 |
| 3n |  | H | 2-CH3,4-Cl | C3H5N2 | H | 4.61 |
| 4a |  | NH2 | 3-Cl | C3H5N2 | CH3 | <4 |
| 4b |  | NH2 | 3,5-diCl | C3H5N2 | CH3 | 4.04 |
| 4c |  | NH2 | 3,4-diCl | C3H5N2 | CH3 | 4.03 |
| 5a |  | NH2 | 3-Cl | C4H7N2 | H | <4 |
| 5b |  | NH2 | 3,5-diCl | C4H7N2 | H | 4.48 |
| 5c |  | NH2 | 3,4-diCl | C4H7N2 | H | 4.46 |
| Bz |  | - | - | - | 5.33 | |
| Properties | Compounds Result (Probability %) | |||
|---|---|---|---|---|
| Bz | 3g | 3j | 3m | |
| Absorption | ||||
| Human Intestinal Absorption | +0.95 | +0.99 | +0.99 | +1.00 |
| Caco-2 | +0.74 | +0.89 | +0.83 | +0.82 |
| Blood-brain barrier | +0.98 | +0.99 | +0.99 | +0.99 |
| Human oral bioavailability | +0.73 | +0.87 | +0.84 | +0.83 |
| Distribution | ||||
| Subcellular localization | Mitochondria 0.77 | Mitochondria 0.38 | Lysosome 0.40 | Mitochondria 0.47 |
| Metabolism | ||||
| P-glycoprotein inhibitor | −0.96 | −0.94 | −0.95 | −0.95 |
| P-glycoprotein substrate | −0.80 | −0.70 | −0.63 | +0.61 |
| CYP3A4 substrate | −0.51 | +0.55 | −0.53 | +0.58 |
| CYP2C9 substrate | −0.78 | −1.00 | −1.00 | −0.79 |
| CYP2D6 substrate | −0.91 | −0.90 | −0.89 | −0.91 |
| CYP3A4 inhibition | −0.82 | −0.93 | −0.88 | −0.88 |
| CYP2C9 inhibition | −0.90 | −0.67 | −0.66 | −0.66 |
| CYP2C19 inhibition | −0.82 | −0.69 | −0.71 | −0.59 |
| CYP2D6 inhibition | −0.92 | −0.82 | −0.85 | −0.75 |
| CYP1A2 inhibition | −0.80 | +0.88 | +0.88 | +0.88 |
| CYP inhibitory promiscuity | −0.58 | −0.66 | −0.56 | −0.54 |
| Toxicity | ||||
| Carcinogenicity (binary) | −0.83 | −0.89 | −0.89 | −0.87 |
| Carcinogenicity (trinary) | Warning 0.41 | No 0.50 | No 0.48 | No 0.55 |
| Ames mutagenesis | +0.93 | −0.79 | −0.77 | −0.80 |
| hERG inhibition | −0.65 | +0.76 | +0.76 | +0.81 |
| Hepatotoxicity | +0.73 | +0.88 | +0.65 | +0.63 |
| Acute Oral Toxicity | III 0.60 | III 0.59 | III 0.60 | III 0.58 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Orlando, L.M.R.; Lechuga, G.C.; da Silva Lara, L.; Ferreira, B.S.; Pereira, C.N.; Silva, R.C.; dos Santos, M.S.; Pereira, M.C.S. Structural Optimization and Biological Activity of Pyrazole Derivatives: Virtual Computational Analysis, Recovery Assay and 3D Culture Model as Potential Predictive Tools of Effectiveness against Trypanosoma cruzi. Molecules 2021, 26, 6742. https://doi.org/10.3390/molecules26216742
Orlando LMR, Lechuga GC, da Silva Lara L, Ferreira BS, Pereira CN, Silva RC, dos Santos MS, Pereira MCS. Structural Optimization and Biological Activity of Pyrazole Derivatives: Virtual Computational Analysis, Recovery Assay and 3D Culture Model as Potential Predictive Tools of Effectiveness against Trypanosoma cruzi. Molecules. 2021; 26(21):6742. https://doi.org/10.3390/molecules26216742
Chicago/Turabian StyleOrlando, Lorraine Martins Rocha, Guilherme Curty Lechuga, Leonardo da Silva Lara, Byanca Silva Ferreira, Cynthia Nathalia Pereira, Rafaela Corrêa Silva, Maurício Silva dos Santos, and Mirian Claudia S. Pereira. 2021. "Structural Optimization and Biological Activity of Pyrazole Derivatives: Virtual Computational Analysis, Recovery Assay and 3D Culture Model as Potential Predictive Tools of Effectiveness against Trypanosoma cruzi" Molecules 26, no. 21: 6742. https://doi.org/10.3390/molecules26216742
APA StyleOrlando, L. M. R., Lechuga, G. C., da Silva Lara, L., Ferreira, B. S., Pereira, C. N., Silva, R. C., dos Santos, M. S., & Pereira, M. C. S. (2021). Structural Optimization and Biological Activity of Pyrazole Derivatives: Virtual Computational Analysis, Recovery Assay and 3D Culture Model as Potential Predictive Tools of Effectiveness against Trypanosoma cruzi. Molecules, 26(21), 6742. https://doi.org/10.3390/molecules26216742