The Influence of 5′,8-Cyclo-2′-deoxypurines on the Mitochondrial Repair of Clustered DNA Damage in Xrs5 Cells: The Preliminary Study
Abstract
:1. Introduction
2. Results and Discussion
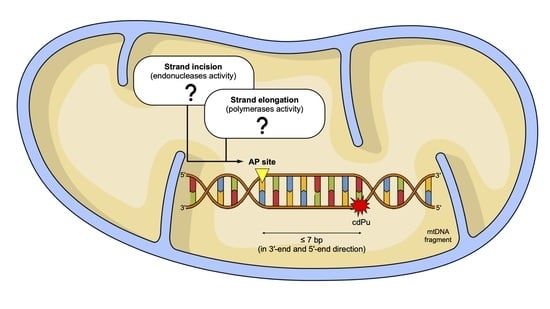
2.1. The Influence of 5′,8-Cyclo-2′-deoxypurines on the Endonucleolytic Activity in Mitochondria
2.2. The Influence of 5′,8-Cyclo-2′-deoxypurines on DNA Synthesis in Mitochondria
3. Materials and Methods
3.1. The Substrate Oligonucleotides
3.2. Preparation of the Mitochondrial Extracts
3.3. Repair Assays
4. Conclusions
- In this study, it has been shown for the first time the efficiency of the initial mtBER steps depends on the distance between AP site and cdPus within bi-stranded CDL, and the type and diastereomeric form of the cdPu.
- In all cases, mitochondrial strand incision and gap-filling were detected for AP sites accompanied by cdPus within bi-stranded CDL.
- The strand incision step of mtBER was enhanced in the presence of cdPu within the cluster compared to Control 1 (single AP site).
- AP site incision was more efficient when AP site was accompanied by RcdA or ScdG than by ScdA or RcdG.
- The gap-filling step of mtBER was inhibited for AP sites located on the 5′-end side of cdPus while for AP sites located on the 3′-end side of cdPus was enhanced compared to Control 1 (single AP site).
- AP sites located in positions denoted as dU0 and dU+1 inhibit endonuclease and polymerases activity in ME, which aligns with previous observations for NE [20].
- Both investigated mtBER stages (strand incision and elongation) showed lower efficiency for AP sites located on the 5′-end side of cdPus, compared to those on the 3′-end side of cdPus. It may be assumed that mtBER is slowed down for AP sites located on the 5′-end side of cdPus.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Boguszewska, K.; Szewczuk, M.; Kaźmierczak-Barańska, J.; Karwowski, B.T. The similarities between human mitochondria and bacteria in the context of structure, genome, and base excision repair system. Molecules 2020, 25, 2857. [Google Scholar] [CrossRef]
- Garcia, I.; Jones, E.; Ramos, M.; Innis-Whitehouse, W.; Gilkerson, R. The little big genome: The organization of mitochondrial DNA. Front. Biosci. (Landmark) 2017, 22, 710–721. [Google Scholar]
- Wasilewski, M.; Chojnacka, K.; Chacinska, A. Protein trafficking at the crossroads to mitochondria. Biochim. Biophys. Acta—Mol. Cell Res. 2017, 1864, 125–137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Topf, U.; Uszczynska-Ratajczak, B.; Chacinska, A. Mitochondrial stress-dependent regulation of cellular protein synthesis. J. Cell Sci. 2019, 132, jcs226258. [Google Scholar] [CrossRef] [Green Version]
- Sharma, P.; Sampath, H. Mitochondrial DNA integrity: Role in health and disease. Cells 2019, 8, 100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- López-Lluch, G.; Santos-Ocaña, C.; Sánchez-Alcázar, J.A.; Fernández-Ayala, D.J.M.; Asencio-Salcedo, C.; Rodríguez-Aguilera, J.C.; Navas, P. Mitochondrial responsibility in ageing process: Innocent, suspect or guilty. Biogerontology 2015, 16, 599–620. [Google Scholar] [CrossRef]
- Martín Giménez, V.M.; de las Heras, N.; Ferder, L.; Lahera, V.; Reiter, R.J.; Manucha, W. Potential effects of melatonin and micronutrients on mitochondrial dysfunction during a cytokine storm typical of oxidative/inflammatory diseases. Diseases 2021, 9, 30. [Google Scholar] [CrossRef] [PubMed]
- Bader, G.; Enkler, L.; Araiso, Y.; Hemmerle, M.; Binko, K.; Baranowska, E.; de Craene, J.O.; Ruer-Laventie, J.; Pieters, J.; Tribouillard-Tanvier, D.; et al. Assigning mitochondrial localization of dual localized proteins using a yeast bi-genomic mitochondrial-split-Gfp. eLife 2020, 9, e56649. [Google Scholar] [CrossRef]
- Lindahl, T. Instability and decay of the primary structure of DNA. Nature 1993, 362, 709–715. [Google Scholar] [CrossRef]
- Karwowski, B.T.; Bellon, S.; O’Neill, P.; Lomax, M.E.; Cadet, J. Effects of (5′S)-5′,8-Cyclo-2′-deoxyadenosine on the base excision repair of oxidatively generated clustered DNA damage. A Biochemical and theoretical study. Org. Biomol. Chem. 2014, 12, 8671–8682. [Google Scholar] [CrossRef]
- Kuraoka, I.; Bender, C.; Romieu, A.; Cadet, J.; Wood, R.D.; Lindahl, T. Removal of oxygen free-radical-induced 5′,8-purine cyclodeoxynucleosides from DNA by the nucleotide excision-repair pathway in human cells. Proc. Natl. Acad. Sci. USA 2000, 97, 3832–3837. [Google Scholar] [CrossRef] [Green Version]
- Jaruga, P.; Dizdaroglu, M. 8,5′-Cyclopurine-2′-deoxynucleosides in DNA: Mechanisms of formation, measurement, repair and biological effects. DNA Repair 2008, 7, 1413–1425. [Google Scholar] [CrossRef]
- Kusumoto, R.; Masutani, C.; Iwai, S.; Hanaoka, F. Translesion synthesis by human DNA polymerase η across thymine glycol lesions. Biochemistry 2002, 41, 6090–6099. [Google Scholar] [CrossRef] [PubMed]
- Brooks, P.J. The cyclopurine deoxynucleosides: DNA repair, biological effects, mechanistic insights, and unanswered questions. Free Radic. Biol. Med. 2017, 107, 90–100. [Google Scholar] [CrossRef] [PubMed]
- Jasti, V.P.; Das, R.S.; Hilton, B.A.; Weerasooriya, S.; Zou, Y.; Basu, A.K. (5′S)-8,5′-cyclo-2′-deoxyguanosine is a strong block to replication, a potent pol V-dependent mutagenic lesion, and is inefficiently repaired in Escherichia coli. Biochemistry 2011, 50, 3862–3865. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.; Xu, M.; Lai, Y.; Laverde, E.E.; Terzidis, M.A.; Masi, A.; Chatgilialoglu, C.; Liu, Y. Bypass of a 5′,8-cyclopurine-2′-deoxynucleoside by DNA polymerase β during DNA replication and base excision repair leads to nucleotide misinsertions and DNA strand breaks. DNA Repair 2015, 33, 24–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lomax, M.E.; Cunniffe, S.; O’Neill, P. Efficiency of repair of an abasic site within DNA clustered damage sites by mammalian cell nuclear extracts. Biochemistry 2004, 43, 11017–11026. [Google Scholar] [CrossRef]
- David-Cordonniert, M.-H.; Laval, J.; O’Neil, P. Clustered DNA damage, influence on damage excision by XRS5 nuclear extracts and escherichia coli Nth and Fpg proteins. J. Biol. Chem. 2000, 275, 11865–11873. [Google Scholar] [CrossRef] [Green Version]
- Eccles, L.J.; Menoni, H.; Angelov, D.; Lomax, M.E.; O’Neill, P. Efficient cleavage of single and clustered AP site lesions within mono-nucleosome templates by CHO-K1 nuclear extract contrasts with retardation of incision by purified APE1. DNA Repair 2015, 35, 27–36. [Google Scholar] [CrossRef] [Green Version]
- Boguszewska, K.; Szewczuk, M.; Kaźmierczak-Barańska, J.; Karwowski, B.T. How (5′S) and (5′R) 5′,8-cyclo-2′-deoxypurines affect base excision repair of clustered DNA damage in nuclear extracts of Xrs5 cells? A biochemical study. Cells 2021, 10, 725. [Google Scholar] [CrossRef]
- Karwowski, B.T. The influence of (5′R)- and (5′S)-5′,8-cyclo-2′-deoxyadenosine on UDG and HAPE1 activity. Tandem lesions are the base excision repair system’s nightmare. Cells 2019, 8, 1303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Izumi, T.; Mellon, I. Base Excision Repair and Nucleotide Excision Repair; Elsevier: Amsterdam, The Netherlands, 2016; ISBN 9780128033456. [Google Scholar]
- Singatulina, A.S.; Pestryakov, P.E. Mechanisms of DNA repair in mitochondria. Biopolym. Cell 2016, 32, 245–261. [Google Scholar] [CrossRef]
- Alexeyev, M.; Shokolenko, I.; Wilson, G.; LeDoux, S. The maintenance of mitochondrial DNA integrity—Critical analysis and update. Cold Spring Harb. Perspect. Biol. 2013, 5, a012641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Omar García-Lepe, U.; Ma Bermúdez-Cruz, R. Mitochondrial genome maintenance: Damage and repair pathways. In DNA Repair-An Update; IntechOpen: London, UK, 2019. [Google Scholar]
- Karwowski, B.T. (5′S) 5′,8-cyclo-2′-deoxyadenosine cannot stop BER. Clustered DNA lesion studies. Int. J. Mol. Sci. 2021, 22, 5934. [Google Scholar] [CrossRef]
- Jaruga, P.; Rozalski, R.; Jawien, A.; Migdalski, A.; Olinski, R.; Dizdaroglu, M. DNA damage products (5′R)- and (5′S)-8,5′-cyclo- 2′-deoxyadenosines as potential biomarkers in human urine for atherosclerosis. Biochemistry 2012, 51, 1822–1824. [Google Scholar] [CrossRef] [PubMed]
- Kant, M.; Akış, M.; Çalan, M.; Arkan, T.; Bayraktar, F.; Dizdaroglu, M.; İşlekel, H. Elevated urinary levels of 8-oxo-2′-deoxyguanosine, (5′R)- and (5′S)-8,5′-cyclo-2′-deoxyadenosines, and 8-iso-prostaglandin F2α as potential biomarkers of oxidative stress in patients with prediabetes. DNA Repair 2016, 48, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Jaruga, P.; Dizdaroglu, M. Identification and quantification of (5′R)- and (5′S)-8,5′-cyclo-2′-deoxyadenosines in human urine as putative biomarkers of oxidatively induced damage to DNA. Biochem. Biophys. Res. Commun. 2010, 397, 48–52. [Google Scholar] [CrossRef]
- Chacinska, A.; Koehler, C.M.; Milenkovic, D.; Lithgow, T.; Pfanner, N. Importing mitochondrial proteins: Machineries and mechanisms. Cell 2009, 138, 628–644. [Google Scholar] [CrossRef] [Green Version]
- Barchiesi, A.; Bazzani, V.; Tolotto, V.; Elancheliyan, P.; Wasilewski, M.; Chacinska, A.; Vascotto, C. Mitochondrial oxidative stress induces rapid intermembrane space/matrix translocation of apurinic/apyrimidinic endonuclease 1 protein through TIM23 complex. J. Mol. Biol. 2020, 432, 166713. [Google Scholar] [CrossRef] [PubMed]
- Pednekar, V.; Weerasooriya, S.; Jasti, V.P.; Basu, A.K. Mutagenicity and genotoxicity of (5′S)-8,5′-cyclo-2′- deoxyadenosine in Escherichia coli and replication of (5′S)-8,5′-cyclopurine-2′-deoxynucleosides in vitro by DNA polymerase Iv, exo-free Klenow fragment, and Dpo4. Chem. Res. Toxicol. 2014, 27, 200–210. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Lai, Y.; Jiang, Z.; Terzidis, M.A.; Masi, A.; Chatgilialoglu, C.; Liu, Y. A 5′, 8-cyclo-2′-deoxypurine lesion induces trinucleotide repeat deletion via a unique lesion bypass by DNA polymerase β. Nucleic Acids Res. 2014, 42, 13749–13763. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krasich, R.; Copeland, W.C. DNA Polymerases in the mitochondria: A critical review of the evidence. Front. Biosci. 2017, 22, 692–709. [Google Scholar] [CrossRef]
- Baptiste, B.A.; Baringer, S.L.; Kulikowicz, T.; Sommers, J.A.; Croteau, D.L.; Brosh, R.M.; Bohr, V.A. DNA polymerase β outperforms DNA polymerase γ in key mitochondrial base excision repair activities. DNA Repair 2021, 99, 103050. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Boguszewska, K.; Kaźmierczak-Barańska, J.; Karwowski, B.T. The Influence of 5′,8-Cyclo-2′-deoxypurines on the Mitochondrial Repair of Clustered DNA Damage in Xrs5 Cells: The Preliminary Study. Molecules 2021, 26, 7042. https://doi.org/10.3390/molecules26227042
Boguszewska K, Kaźmierczak-Barańska J, Karwowski BT. The Influence of 5′,8-Cyclo-2′-deoxypurines on the Mitochondrial Repair of Clustered DNA Damage in Xrs5 Cells: The Preliminary Study. Molecules. 2021; 26(22):7042. https://doi.org/10.3390/molecules26227042
Chicago/Turabian StyleBoguszewska, Karolina, Julia Kaźmierczak-Barańska, and Bolesław T. Karwowski. 2021. "The Influence of 5′,8-Cyclo-2′-deoxypurines on the Mitochondrial Repair of Clustered DNA Damage in Xrs5 Cells: The Preliminary Study" Molecules 26, no. 22: 7042. https://doi.org/10.3390/molecules26227042
APA StyleBoguszewska, K., Kaźmierczak-Barańska, J., & Karwowski, B. T. (2021). The Influence of 5′,8-Cyclo-2′-deoxypurines on the Mitochondrial Repair of Clustered DNA Damage in Xrs5 Cells: The Preliminary Study. Molecules, 26(22), 7042. https://doi.org/10.3390/molecules26227042