Isolation and Identification of Pentalenolactone Analogs from Streptomyces sp. NRRL S-4
Abstract
:1. Introduction

2. Results
2.1. Pentalenolactone BGC in S-4
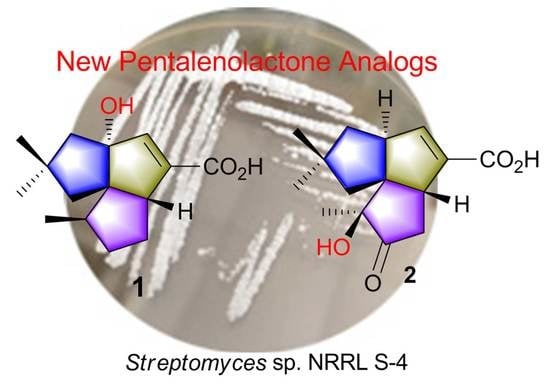
2.2. Isolation and Structural Identification
2.3. Antimicrobial Activity
2.4. Proposed Biosynthetic Pathway
3. Materials and Methods
3.1. Fermentation and Isolation
3.2. General Experimental Procedures
3.3. ECD Calculations
3.4. Antimicrobial Activity Assays
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Musiol-Kroll, E.M.; Tocchetti, A.; Sosio, M.; Stegmann, E. Challenges and advances in genetic manipulation of filamentous actinomycetes—the remarkable producers of specialized metabolites. Nat. Prod. Rep. 2019, 36, 1351–1369. [Google Scholar] [CrossRef] [PubMed]
- Yamada, Y.; Kuzuyama, T.; Komatsu, M.; Shin-Ya, K.; Omura, S.; Cane, D.E.; Ikeda, H. Terpene synthases are widely distributed in bacteria. Proc. Natl. Acad. Sci. USA 2015, 112, 857–862. [Google Scholar] [CrossRef] [Green Version]
- Medema, M.H.; de Rond, T.; Moore, B.S. Mining genomes to illuminate the specialized chemistry of life. Nat. Rev. Genet. 2021, 22, 553–571. [Google Scholar] [CrossRef]
- Geng, W.-L.; Wang, X.-Y.; Kurtán, T.; Mándi, A.; Tang, H.; Schulz, B.; Sun, P.; Zhang, W. Herbarone, a rearranged heptaketide derivative from the sea hare associated fungus Torula herbarum. J. Nat. Prod. 2012, 75, 1828–1832. [Google Scholar] [CrossRef] [PubMed]
- Sun, P.; Xu, D.-X.; Mándi, A.; Kurtán, T.; Li, T.-J.; Schulz, B.; Zhang, W. Structure, absolute configuration, and conformational study of 12-membered macrolides from the fungus Dendrodochium sp. associated with the sea cucumber Holothuria nobilis Selenka. J. Org. Chem. 2013, 78, 7030–7047. [Google Scholar] [CrossRef]
- Li, H.; Zhang, M.; Li, H.; Yu, H.; Chen, S.; Wu, W.; Sun, P. Discovery of venturicidin congeners and identification of the biosynthetic gene cluster from Streptomyces sp. NRRL S-4. J. Nat. Prod. 2021, 84, 110–119. [Google Scholar] [CrossRef] [PubMed]
- Frattaruolo, L.; Lacret, R.; Cappello, A.R.; Truman, A.W. A genomics-based approach identifies a thioviridamide-like compound with selective anticancer activity. ACS Chem. Biol. 2017, 12, 2815–2822. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.H.; Weber, T. antiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019, 47, W81–W87. [Google Scholar] [CrossRef] [Green Version]
- Seo, M.J.; Zhu, D.; Endo, S.; Ikeda, H.; Cane, D.E. Genome mining in Streptomyces. Elucidation of the role of Baeyer-Villiger monooxygenases and non-heme iron-dependent dehydrogenase/oxygenases in the final steps of the biosynthesis of pentalenolactone and neopentalenolactone. Biochemistry 2011, 50, 1739–1754. [Google Scholar] [CrossRef] [Green Version]
- Koe, B.K.; Sobin, B.A.; Celmer, W.D. PA 132, a new antibiotic. I. Isolation and chemical properties. Antibiot. Annu. 1957, 672–675. [Google Scholar]
- Takeuchi, S.; Ogawa, Y.; Yonehara, H. The structure of pentalenolactone (PA-132). Tetrahedron Lett. 1969, 32, 2737–2740. [Google Scholar]
- Martin, D.; Slomp, G.; Mizsak, S.; Duchamp, D.; Chidester, C. The structure and absolute configuration of pentalenolactone (PA 132). Tetrahedron Lett. 1970, 56, 4901–4904. [Google Scholar] [CrossRef]
- Duszenko, M.; Balla, H.; Mecke, D. Specific inactivation of glucose metabolism from eucaryotic cells by pentalenolactone. Biochim. Biophys. Acta 1982, 714, 344–350. [Google Scholar] [CrossRef]
- Quaderer, R.; Omura, S.; Ikeda, H.; Cane, D.E. Pentalenolactone biosynthesis. Molecular cloning and assignment of biochemical function to PtlI, a cytochrome P450 of Streptomyces avermitilis. J. Am. Chem. Soc. 2006, 128, 13036–13037. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tetzlaff, C.N.; You, Z.; Cane, D.E.; Takamatsu, S.; Omura, S.; Ikeda, H. A gene cluster for biosynthesis of the sesquiterpenoid antibiotic pentalenolactone in Streptomyces avermitilis. Biochemistry 2006, 45, 6179–6186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- You, Z.; Omura, S.; Ikeda, H.; Cane, D.E. Pentalenolactone biosynthesis. molecular cloning and assignment of biochemical function to PtlH, a non-heme iron dioxygenase of Streptomyces avermitilis. J. Am. Chem. Soc. 2006, 128, 6566–6567. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duan, L.; Jogl, G.; Cane, D.E. The cytochrome P450-catalyzed oxidative rearrangement in the final step of pentalenolactone biosynthesis: Substrate structure determines mechanism. J. Am. Chem. Soc. 2016, 138, 12678–12689. [Google Scholar] [CrossRef] [Green Version]
- Seto, H.; Sasaki, T.; Yonehara, H.; Takahashi, S.; Takeuchi, M.; Kuwano, H.; Arai, M. Studies on the biosynthesis of pentalenolactone. VII. Isolation of pentalenolactones P and O. J. Antibiot. 1984, 37, 1076–1078. [Google Scholar] [CrossRef] [Green Version]
- Cane, D.E.; Sohng, J.K.; Williard, P.G. Isolation and structure determination of pentalenolactones A, B, D, and F. J. Org. Chem. 1992, 57, 844–851. [Google Scholar] [CrossRef]
- Takamatsu, S.; Xu, L.H.; Fushinobu, S.; Shoun, H.; Komatsu, M.; Cane, D.E.; Ikeda, H. Pentalenic acid is a shunt metabolite in the biosynthesis of the pentalenolactone family of metabolites: Hydroxylation of 1-deoxypentalenic acid mediated by CYP105D7 (SAV_7469) of Streptomyces avermitilis. J. Antibiot. 2011, 64, 65–71. [Google Scholar] [CrossRef] [Green Version]
- Rebehmed, J.; Alphand, V.; de Berardinis, V.; de Brevern, A.G. Evolution study of the Baeyer-Villiger monooxygenases enzyme family: Functional importance of the highly conserved residues. Biochimie 2013, 95, 1394–1402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A.; et al. Gaussian 09, Revision B.01; Gaussian, Inc.: Wallingford, CT, USA, 2010. [Google Scholar]
- MacroModel 10.2; Schrödinger, LLC: New York, NY, USA, 2009.





| Protein | Size (aa) | Protein Homologue and Origin | Identity/Similarity, %/% | Proposed Function |
|---|---|---|---|---|
| PllN | 334 | GapR, S. arenae TU469 | 93/100 | glyceraldehyde-3-phosphate dehydrogenase |
| PllM | 398 | PntM, S. arenae TU469 | 87/100 | cytochrome P450 monooxygenase |
| PllH | 283 | PntH, S. arenae TU469 | 89/100 | 1-deoxypentalenic acid-11-beta-hydroxylas |
| PllG | 484 | PntG, S. arenae TU469 | 84/99 | transmembrane efflux protein |
| PllF | 275 | PntF, S. arenae TU469 | 91/91 | short-chain dehydrogenase/reductase |
| PllE | 584 | PntE, S. arenae TU469 | 91/100 | Baeyer-Villiger monooxygenase |
| PllD | 299 | PntD, S. arenae TU469 | 89/100 | non-heme iron/alpha-ketoglutarate-dependent dioxygenase |
| PllB | 337 | PntB, S. arenae TU469 | 93/99 | polyprenyl diphosphate synthase |
| PllA | 337 | PntA, S. arenae TU469 | 100/100 | pentalenene synthease |
| PllI | 457 | PntI, S. arenae TU469 | 92/97 | cytochrome P450 monooxygenase |
| 1 | 2 | ||||
|---|---|---|---|---|---|
| no. | δC, Type | δH (J in Hz) | δC, Type | δH (J in Hz) | |
| 1α | 55.3, CH2 | 1.70, d (13.5) | 1α | 46.2, CH2 | 1.68, m |
| 1β | 1.87, dd (13.5, 1.3) | 1β | 1.68, m | ||
| 2 | 38.1, C | 2 | 40.2, C | ||
| 3α | 50.1, CH2 | 1.94, d (13.2) | 3α | 49.7, CH2 | 2.25, d (14.7) |
| 3β | 1.50, dd (13.2, 1.3) | 3β | 1.59, d (14.7) | ||
| 4 | 65.4, C | 4 | 65.9, C | ||
| 5 | 60.5, CH | 3.02, dd (8.4, 4.4) | 5 | 51.9, CH | 3.37, brd (11.2) |
| 6 | 140.1, C | 6 | 139.8, C | ||
| 7 | 148.0, CH | 6.41, s | 7 | 149.8, CH | 6.62, brd (2.0) |
| 8 | 94.4, C | 8 | 55.3, CH | 2.94, ddd (7.7, 2.5, 2.0) | |
| 9 | 39.2, CH | 2.25, m | 9 | 80.5, C | |
| 10 | 17.0, CH3 | 0.96, d (7.1) | 10 | 17.1, CH3 | 1.18, s |
| 11α | 35.1, CH2 | 1.72, m | 11 | 217.1, C | |
| 11β | 1.28, dq (12.3, 6.3) | ||||
| 12α | 31.1, CH2 | 1.44, ddd (18.9, 6.3, 4.4) | 12α | 41.2, CH2 | 1.91, dd (18.5, 4.2) |
| 12β | 2.02, m | 12β | 3.07, dd (18.5, 11.2) | ||
| 13 | 169.2, C | 13 | 167.9, C | ||
| 14 | 27.7, CH3 | 0.94, s | 14 | 31.9, CH3 | 1.09, s |
| 15 | 32.1, CH3 | 1.06, s | 15 | 30.9, CH3 | 1.04, s |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, H.; Li, H.; Chen, S.; Wu, W.; Sun, P. Isolation and Identification of Pentalenolactone Analogs from Streptomyces sp. NRRL S-4. Molecules 2021, 26, 7377. https://doi.org/10.3390/molecules26237377
Li H, Li H, Chen S, Wu W, Sun P. Isolation and Identification of Pentalenolactone Analogs from Streptomyces sp. NRRL S-4. Molecules. 2021; 26(23):7377. https://doi.org/10.3390/molecules26237377
Chicago/Turabian StyleLi, Huanhuan, Hongji Li, Shuo Chen, Wenhui Wu, and Peng Sun. 2021. "Isolation and Identification of Pentalenolactone Analogs from Streptomyces sp. NRRL S-4" Molecules 26, no. 23: 7377. https://doi.org/10.3390/molecules26237377
APA StyleLi, H., Li, H., Chen, S., Wu, W., & Sun, P. (2021). Isolation and Identification of Pentalenolactone Analogs from Streptomyces sp. NRRL S-4. Molecules, 26(23), 7377. https://doi.org/10.3390/molecules26237377