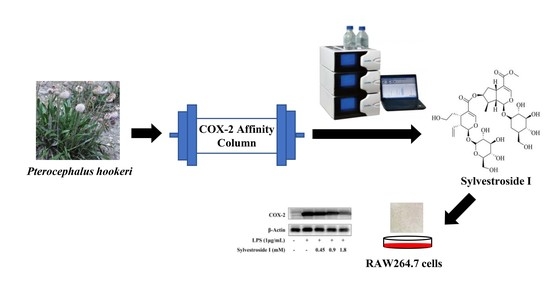
Targeted Separation of COX-2 Inhibitor from Pterocephalus hookeri Using Preparative High-Performance Liquid Chromatography Directed by the Affinity Solid-Phase Extraction HPLC System
Abstract
:1. Introduction
2. Materials and Methods
2.1. Instrumentation and Reagents
2.2. Sample Extraction and Pretreatment
2.3. Preparation of Affinity Solid-Phase Extraction Column
2.4. Chromatographic Conditions
2.5. Determination of Anti-Inflammatory Activity
2.5.1. COX-2 Inhibitory Activities Assay
2.5.2. Cell Culture and Treatment
2.5.3. Cell Viability Assay
2.5.4. PGE2 Levels Analysis
2.5.5. COX-2 Levels Analysis
2.5.6. Molecular Docking
3. Results and Discussion
3.1. P. hookeri Extract Pretreatment by Medium-Pressure Chromatography
3.2. Identification of COX-2 Inhibitors with the ASPE-HPLC System
3.3. Enrichment of COX-2 Inhibitors Fraction
3.4. Purification and Characterization of COX-2 Inhibitor
3.5. In Vitro COX-2 Inhibitory Activity Assay
4. Concluding Remarks
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
Abbreviations
References
- Li, L.Y.; Ciren, B.Z.; Zhan, D.; Wei, Y.F. Comprehensive utilization and development of traditional Tibetan medicine in China. Chin. J. Chin. Mater. Med. 2001, 26, 808–810. [Google Scholar]
- Dong, Z.Y.; Wei, L.; Lu, H.Q.; Zeng, Q.H.; Meng, F.C.; Wang, G.W.; Lan, X.Z.; Liao, Z.H.; Chen, M. Ptehoosines A and B: Two new sesamin-type sesquilignans with antiangiogenic activity from Pterocephalus hookeri (C.B. Clarke) Heck. Fitoterapia 2021, 151, 104886. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.C.; Ying, Y.J.; Guo, F.J.; Zhu, G.F. Bis-iridoid and lignans from traditional Tibetan herb Pterocephalus hookeri. Biochem. Syst. Ecol. 2014, 56, 209–212. [Google Scholar] [CrossRef]
- Yang, P.; Li, Y.Q.; Liu, X.; Jiang, S.X. Determination of free isomeric oleanolic acid and ursolic acid in Pterocephalus hookeri by capillary zone electrophoresis. J. Pharm. Biomed. Anal. 2007, 43, 1331–1334. [Google Scholar] [CrossRef]
- Wang, W.X.; Luo, S.Y.; Wang, Y.; Xiang, L.; Liu, X.H.; Tang, C.; Zhang, Y. Pterocephanoside A, a new iridoid from a traditional Tibetan medicine, Pterocephalus hookeri. J. Asian Nat. Prod. Res. 2020, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Hu, J.J.; Lin, J.W.; Fang, W.S.; Du, G.H. Anti-inflammatory and analgesic effects of ethanol and aqueous extracts of Pterocephalus hookeri (C.B. Clarke) Höeck. J. Ethnopharmacol. 2009, 123, 510–514. [Google Scholar] [CrossRef]
- Guo, C.X.; Wu, Y.C.; Zhu, Y.Z.; Wang, Y.C.; Tian, L.L.; Lu, Y.; Han, C.; Zhu, G.F. In Vitro and In Vivo Antitumor Effects of n-Butanol Extracts of Pterocephalus hookeri on Hep3B Cancer Cell. Evid. Based Complement. Altern. Med. 2015, 2015, 159132. [Google Scholar] [CrossRef] [Green Version]
- Guo, C.X.; Wu, Y.C.; Zhu, Y.Z.; Tian, L.L.; Lu, Y.; Han, C.; Zhu, G.F. Inhibits Human Liver Hep3B Cell Proliferation and Invasion and Mestasis of n-butanol Part of Pterocephalus hookeri in Vitro. Chin. J. Exp. Tradit. Med. Form. 2015, 21, 100–105. [Google Scholar]
- Yang, P.F.; Lu, H.; Wang, Q.B.; Zhao, Z.W.; Liu, Q.; Zhao, X.; Yang, J.; Huang, S.; Chen, Z.F.; Mao, D.B. Chemical Composition and Antimicrobial Activities of the Essential Oil from the Leaves of Pterocephalus hookeri. Nat. Prod. Commun. 2020, 15, 1934578X20981239. [Google Scholar] [CrossRef]
- LI, G.Q.; Sheng, D.L. Chemical constituents from Pterocephalus hookeri and their neuroprotection activities. Chin. Tradit. Pat. Med. 2018, 40, 1329–1335. [Google Scholar]
- Tang, C.; Li, H.J.; Fan, G.; Kuang, T.T.; Meng, X.L.; Zou, Z.M.; Zhang, Y. Network pharmacology and UPLC-Q-TOF/MS studies on the anti-arthritic mechanism of Pterocephalus hookeri. Trop. J. Pharm. Res. 2018, 17, 1095–1110. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.C.; Guo, C.X.; Zhu, Y.Z.; Li, Y.M.; Guo, F.J.; Zhu, G.F. Four new bis-iridoids isolated from the traditional Tibetan herb Pterocephalus hookeri. Fitooterapia. 2014, 98, 104–109. [Google Scholar] [CrossRef]
- Wu, Y.C.; Lu, J.; Lu, X.Q.Y.; Li, R.; Guo, J.; Guo, F.J.; Li, Y.M. Monoterpenoids and Triterpenoids from Pterocephalus hookeri with NF-κB inhibitory activity. Phytochem. Lett. 2015, 13, 30–34. [Google Scholar] [CrossRef]
- Chen, Y.L.; Yu, H.; Guo, F.J.; Wu, Y.C.; Li, Y.M. Antinociceptive and anti-inflammatory activities of a standardized extract of bis-iridoids from Pterocephalus hookeri. J. Ethnopharmacol. 2018, 216, 233–238. [Google Scholar] [CrossRef]
- Zhu, Y.H.; Wang, W.D.; Liu, Z.G.; Jiang, S.R.; Tao, Y.D.; Jiang, L.; Mei, L.J. Comprehensive screening and separation of cyclooxygenase-2 inhibitors from Pterocephalus hookeri by affinity solid-phase extraction coupled with preparative high-performance liquid chromatography. J. Chromatogr. B 2021, 1183, 122981. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Dong, Z.Y.; Lan, X.Z.; Liao, Z.H.; Chen, M. Sweroside Alleviated LPS-Induced Inflammation via SIRT1 Mediating NF-κB and FOXO1 Signaling Pathways in RAW264.7 Cells. Molecules 2019, 24, 872. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gashaw, I.; Ellinghaus, P.; Sommer, A.; Asadullah, K. What makes a good drug target? Drug Discov. Today 2012, 17, S24–S30. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.F.; He, R.J.; Hou, X.D.; Ji, D.R.; Zhang, Q.H.; Wang, P.; Ge, G.B. New technologies for effient discovery and evaluation of natural enzyme inhibitors: Research progress and perspectives. Acad. J. Shanghai Univ. Tradit. Chin. Med. 2021, 35, 1-11+19. [Google Scholar]
- Gedawy, E.M.; Kassab, A.E.; Kerdawy, A.M.E. Design, synthesis and biological evaluation of novel pyrazole sulfonamide derivatives as dual COX-2/5-LOX inhibitors. Eur. J. Med. Chem. 2020, 189, 112066. [Google Scholar] [CrossRef]
- Fitzpatrick, F.A. Cyclooxygenase enzymes: Regulation and function. Curr. Pharm. Design 2004, 10, 577–588. [Google Scholar] [CrossRef]
- Attiq, A.; Jalil, J.; Husain, K.; Ahmad, W. Raging the War against Inflammation with Natural Products. Front. Pharmacol. 2018, 9, 976. [Google Scholar] [CrossRef] [PubMed]
- Sobolewski, C.; Legrand, N. Celecoxib Analogues for Cancer Treatment: An Update on OSU-03012 and 2,5-Dimethyl-Celecoxib. Biomolecules 2021, 11, 1049. [Google Scholar] [CrossRef]
- Cryer, B. Nonsteroidal anti-inflammatory drug gastrointestinal toxicity. Curr. Opin. Gastroenterol. 2001, 17, 503–512. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.Q.; Guo, L.P.; Liu, C.M.; Li, S.N. Single-step screening and isolation of potential lipoxidase inhibitors from Trifolium repens by stepwise flow rate high-speed countercurrent chromatography and semipreparative high-performance liquid chromatography target-guided by ultrafiltration-LC-MS. J. Sep. Sci. 2021. [CrossRef]
- Peng, M.J.; Shi, S.Y.; Chen, L.; Zhang, S.H.; Cai, P.; Chen, X.Q. Online coupling solid-phase ligand-fishing with high-performance liquid chromatography-diode array detector-tandem mass spectrometry for rapid screening and identification of xanthine oxidase inhibitors in natural products. Anal. Bioanal. Chem. 2016, 408, 6693–6701. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.P.; Shi, S.Y.; Chen, L.; Chen, X.Q.; Zhang, S.H. Rapid screening and identification of α-glucosidase binding compounds in Chinese medicines by online affinity solid-phase extraction-high performance liquid chromatography-diode detector-quadrupole time-of-flight mass spectrometry. Chin. J. Chromatogr. 2017, 35, 274–279. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.H.; Zhao, Y.M.; Zhang, Y.Y.; Zhang, T.T.; Kool, J.; Somsen, G.W.; Wang, Q.Q.; Jiang, Z.J. Online screening of acetylcholinesterase inhibitors in natural products using monolith-based immobilized capillary enzyme reactors combined with liquid chromatography-mass spectrometry. J. Chromatogr. A. 2018, 1563, 135–143. [Google Scholar] [CrossRef]
- Wang, W.D.; Dang, J.; Shao, Y.; Jiang, L.; Liu, Z.G.; Mei, L.J.; Tao, Y.D. A novel chromatographic separation method for rapid enrichment and isolation of novel flavonoid glycosides from Sphaerophysa salsula. J. Sep. Sci. 2020, 43, 4018–4027. [Google Scholar] [CrossRef]
- Dang, J.; Du, Y.R.; Wang, Q.; Dawa, Y.Z.; Chen, C.B.; Wang, Q.L.; Ma, J.B.; Tao, Y.D. Preparative isolation of arylbutanoid-type phenol [(-)-rhododendrin] with peak tailing on conventional C18 column using middle chromatogram isolated gel column coupled with reversed-phase liquid chromatography. J. Sep. Sci. 2020, 43, 3233–3241. [Google Scholar] [CrossRef]
- Fan, Y.P.; Fu, Y.H.; Fu, Q.; Cai, J.F.; Xin, H.X.; Dai, M.; Jin, Y. Purification of flavonoids from licorice using an off-line preparative two-dimensional normal-phase liquid chromatography/reversed-phase liquid chromatography method. J. Sep. Sci. 2016, 39, 2710–2719. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.D.; Tao, Y.D.; Jiao, L.J.; Fan, M.X.; Shao, Y.; Wang, Q.L.; Mei, L.J.; Dang, J. Efficient separation of high-purity compounds from Oxytropis falcata using two-dimensional preparative chromatography. J. Sep. Sci. 2017, 40, 3593–3601. [Google Scholar] [CrossRef] [PubMed]
- Lazaridi, E.; Janssen, H.G.; Vincken, J.P.; Pirok, B.; Hennebelle, M. A comprehensive two-dimensional liquid chromatography method for the simultaneous separation of lipid species and their oxidation products. J. Chromatogr. A 2021, 1644, 462106. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Wang, N.N.; Shen, C.; Li, H.M.; Zhao, J.Y.; Chen, T.; Li, Y.L. An effective method based on medium-pressure liquid chromatography and recycling high-speed counter-current chromatography for enrichment and separation of three minor components with similar polarity from Dracocephalum tanguticum. J. Sep. Sci. 2019, 42, 684–690. [Google Scholar] [CrossRef]
- Qing, L.S.; Xue, Y.; Zheng, Y.; Xiong, J.; Liao, X.; Ding, L.S.; Li, B.G.; Liu, Y.M. Ligand fishing from Dioscorea nipponica extract using human serum albumin functionalized magnetic nanoparticles. J. Chromatogr. A 2021, 1217, 4663–4668. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chan, H.S. Matching speed and locality. Nature 1998, 392, 761–763. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, X.C.; Zhang, Y.L.; Chen, G.L.; Liu, Y.; Hu, X.L.; Li, N.; Wu, J.L.; Guo, M.Q. Identification of Anti-Inflammatory and Anti-Proliferative Neolignanamides from Warburgia ugandensis Employing Multi-Target Affinity Ultrafiltration and LC-MS. Pharmaceuticals 2021, 14, 313. [Google Scholar] [CrossRef]
- Schellinger, A.P.; Carr, P.W. Isocratic and gradient elution chromatography: A comparison in terms of speed, retention reproducibility and quantitation. J. Chromatogr. A 2006, 1109, 253–266. [Google Scholar] [CrossRef]
- Wang, L.Q.; Lu, S.Q.; Wang, L.Y.; Xin, M.; Xu, Y.Y.; Wang, G.; Chen, D.Q.; Chen, L.X.; Liu, S.; Zhao, F. Anti-inflammatory effects of three withanolides isolated from Physalis angulata L. in LPS-activated RAW 264.7 cells through blocking NF-kappa B signaling pathway. J. Ethnopharmacol. 2021, 276, 114186. [Google Scholar] [CrossRef]








Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, Y.; Wang, W.; Jiang, L.; Tan, H.; Liu, Z.; Jiang, S.; Tao, Y.; Wen, H.; Mei, L. Targeted Separation of COX-2 Inhibitor from Pterocephalus hookeri Using Preparative High-Performance Liquid Chromatography Directed by the Affinity Solid-Phase Extraction HPLC System. Molecules 2021, 26, 7395. https://doi.org/10.3390/molecules26237395
Zhu Y, Wang W, Jiang L, Tan H, Liu Z, Jiang S, Tao Y, Wen H, Mei L. Targeted Separation of COX-2 Inhibitor from Pterocephalus hookeri Using Preparative High-Performance Liquid Chromatography Directed by the Affinity Solid-Phase Extraction HPLC System. Molecules. 2021; 26(23):7395. https://doi.org/10.3390/molecules26237395
Chicago/Turabian StyleZhu, Yunhe, Weidong Wang, Lei Jiang, Hui Tan, Zenggen Liu, Sirong Jiang, Yanduo Tao, Huaixiu Wen, and Lijuan Mei. 2021. "Targeted Separation of COX-2 Inhibitor from Pterocephalus hookeri Using Preparative High-Performance Liquid Chromatography Directed by the Affinity Solid-Phase Extraction HPLC System" Molecules 26, no. 23: 7395. https://doi.org/10.3390/molecules26237395
APA StyleZhu, Y., Wang, W., Jiang, L., Tan, H., Liu, Z., Jiang, S., Tao, Y., Wen, H., & Mei, L. (2021). Targeted Separation of COX-2 Inhibitor from Pterocephalus hookeri Using Preparative High-Performance Liquid Chromatography Directed by the Affinity Solid-Phase Extraction HPLC System. Molecules, 26(23), 7395. https://doi.org/10.3390/molecules26237395