Scalarane Sesterterpenoids with Antibacterial and Anti-Proliferative Activities from the Mushroom Neonothopanus nambi
Abstract
:1. Introduction
2. Materials and Methods
2.1. General Experimental Procedure
2.2. Fungus Material
2.3. Extraction and Isolation
2.4. ECD Calculations
2.5. Antibacterial Assay
2.6. Antiproliferative Activity Assay
3. Results
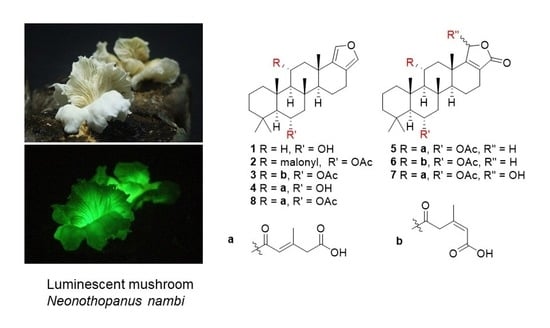
3.1. Isolated Compounds from N. nambi
3.2. Structural Characterization of the New Compounds
3.3. Cytotoxic Activities of Scalarane Sesterterpenoids 1–8
3.4. Antibacterial Activities of Scalarane Sesterterpenoids 1–8
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sangsopha, W.; Lekphrom, R.; Schevenels, F.; Saksirirat, W.; Bua-Art, S.; Kanokmedhakul, K.; Kanokmedhakul, S. New p-terphenyl and benzoquinone metabolites from the bioluminescent mushroom Neonothopanus nambi. Nat. Prod. Res. 2019, 34, 2186–2193. [Google Scholar] [CrossRef] [PubMed]
- Namanusart, W.; Saksirirat, W.; Tanthanuch, W. FTIR spectroscopic analysis of luminescent mushroom (Neonothopanus nambi Speg.) induced disease resistance against tomato root-knot nematode, Meloidogyne incognita Chitwood. J. Pure Appl. Microbiol. 2016, 10, 923–935. [Google Scholar]
- Bua-art, S.; Saksirirat, W.; Kanokmedhakul, S.; Hiransalee, A.; Lekphrom, R. Extraction of bioactive compounds from luminescent mushroom (Neonothopanus nambi) and its effect on root-knot nematode (Meloidogyne incognita). Asia-Pac. J. Sci. Technol. 2010, 15, 726–737. [Google Scholar]
- Kanokmedhakul, S.; Lekphrom, R.; Kanokmedhakul, K.; Hahnvajanawong, C.; Bua-Art, S.; Saksirirat, W.; Prabpai, S.; Kongsaeree, P. Cytotoxic sesquiterpenes from luminescent mushroom Neonothopanus nambi. Tetrahedron 2012, 68, 8261–8266. [Google Scholar] [CrossRef]
- Tsarkova, A.S.; Dubinnyi, M.A.; Baranov, M.S.; Oguienko, A.D.; Yampolsky, I.V. Nambiscalarane, a novel sesterterpenoid comprising a furan ring, and other secondary metabolites from bioluminescent fungus Neonothopanus nambi. Mendeleev Commun. 2016, 26, 191–192. [Google Scholar] [CrossRef]
- Fattorusso, E.; Magno, S.; Santacroce, C.; Sica, D. Scalarin, a new pentacyclic C-25 terpenoid from the sponge Cacospongia scalaris. Tetrahedron 1972, 28, 5993–5997. [Google Scholar] [CrossRef]
- Cimino, G.; De Rosa, S.; De Stefano, S.; Sodano, G. The chemical defense of four Mediterranean nudibranchs. Comp. Biochem. Physiol. Part B Comp. Biochem. 1982, 73, 471–474. [Google Scholar] [CrossRef]
- Doi, Y.; Shigemori, H.; Ishibashi, M.; Mizobe, F.; Kawashima, A.; Nakaike, S.; Kobayashi, J. New Sesterterpenes with Nerve Growth Factor Synthesis-Stimulating Activity from the Okinawan Marine Sponge Hyrtios sp. Chem. Pharm. Bull. 1993, 41, 2190–2191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, X.C.; Liu, H.L.; Lou, L.G.; Guo, Y.W. Chemical constituents of two marine sponges Dysidea villosa and Dysidea marshalla. Chin. J. Nat. Med. 2008, 6, 425–429. [Google Scholar] [CrossRef]
- Mahidol, C.; Prawat, H.; Sangpetsiripan, S.; Ruchirawat, S. Bioactive Scalaranes from the Thai Sponge Hyrtios gumminae. J. Nat. Prod. 2009, 72, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Rogers, S.D.; Paul, V.J. Chemical defenses of three Glossodoris nudibranchs and their dietary Hyrtios sponges. Mar. Ecol. Prog. Ser. 1991, 77, 221–232. [Google Scholar] [CrossRef]
- Shen, X.; Guo, Y.; Jian, H.; Yu, Z.; Du, L. Use of Compound Scalarafuran. CN Patent 1864691 A, 16 June 2010. [Google Scholar]
- Song, J.; Jeong, W.; Wang, N.; Lee, H.-S.; Sim, C.J.; Oh, K.-B.; Shin, J. Scalarane Sesterterpenes from the Sponge Smenospongia sp. J. Nat. Prod. 2008, 71, 1866–1871. [Google Scholar] [CrossRef]
- Zeng, L.M.; Fu, X.; Su, J.Y.; Chen, S.; Snyder, J.K. Phyllofenone A, a new scalarane sesterterpene from the sponge Phyllospongia foliascens (Pallas). Chem. Res. Chin. Univ. 1991, 7, 100–106. [Google Scholar]
- Wang, Q.; Sun, Y.; Yang, L.; Luo, X.; de Voogd, N.J.; Tang, X.; Li, P.; Li, G. Bishomoscalarane sesterterpenoids from the sponge Dysidea granulosa collected in the South China Sea. J. Nat. Prod. 2020, 83, 516–523. [Google Scholar] [CrossRef] [PubMed]
- Hassan, M.H.A.; Rateb, M.E.; Hetta, M.; Abdelaziz, T.A.; Sleim, M.A.; Jaspars, M.; Mohammed, R. Scalarane sesterterpenes from the Egyptian Red Sea sponge Phyllospongia lamellosa. Tetrahedron 2015, 71, 577–583. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A.; et al. Gaussian 09; Revision B.01; Gaussian, Inc.: Wallingford, CT, USA, 2009. [Google Scholar]
- Weinstein, M.P.; Limbago, B.; Patel, J.B.; Mathers, A.J.; Burnham, C.-A.; Mazzulli, T.; Campeau, S.; Munro, S.D.; Conville, P.S.; Ordoñez Smith de Danies, M.; et al. Reference Method for Dilution Antimicrobial Susceptibility Test for Bacteria That Grow Aerobically: Approved Standard M07-A9; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2012. [Google Scholar]
- Kumnerdkhonkaen, P.; Saenglee, S.; Asgar, M.A.; Senawong, G.; Khongsukwiwat, K.; Senawong, T. Antiproliferative activities and phenolic acid content of water and ethanolic extracts of the powdered formula of Houttuynia cordata Thunb. Fermented broth and Phyllanthus emblica Linn. fruit. BMC Complement. Altern. Med. 2018, 18, 130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ungur, N.; Kulcitki, V. Synthetic paths towards scalaranes: Assembling the scalaranic skeleton and further transformations. Phytochem. Rev. 2004, 3, 401–415. [Google Scholar] [CrossRef]
- Pettit, G.R.; Cichacz, Z.A.; Tan, R.; Hoard, M.S.; Melody, N.; Pettit, R.K. Antineoplastic Agents. 386. Isolation of Sesterstatins 1–3 from the Marine Sponge Hyrtios erecta. J. Nat. Prod. 1998, 61, 13–16. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.-L.; Zhang, Z.-G.; Li, H.-C.; Deng, W.-P. Concise stereoselective synthesis of marine sesterterpene, 16-deacetoxy-12-epi-scalarafuran acetate and its 14-epimer via intramolecular Diels-Alder addition. Tetrahedron 2011, 67, 6939–6943. [Google Scholar] [CrossRef]






| No. | 1 a | 2 a | 3 a | 4 b | 5 a | 6 a | 7 c |
|---|---|---|---|---|---|---|---|
| 1 | 1.97 (m) 1.67 (m) | 1.84 (dt, 12.8, 5.2) 1.27 (overlap) | 1.93 (m) 1.21 (overlap) | 1.81 (m) 1.20 (m) | 1.91 (m) 1.28 (overlap) | 1.90 (m) 1.22 (overlap) | 1.96 (m) 1.38 (m) |
| 2 | 1.60 (m) 1.42 (m) | 1.53 (m) 1.39 (m) | 1.51 (m) 1.37 (overlap) | 1.45 (m) 1.32 (overlap) | 1.53 (m) 1.40 (m) | 1.54 (m) 1.38 (m) | 1.58 (m) 1.44 (m) |
| 3 | 1.35 (m) 1.20 (m) | 1.37 (overlap) 1.20 (overlap) | 1.37 (overlap) 1.18 (overlap) | 1.31 (overlap) 1.11 (overlap) | 1.37 (m) 1.19 (m) | 1.37 (m) 1.22 (overlap) | 1.38 (m) 1.23 (m) |
| 4 | - | - | - | - | - | - | - |
| 5 | 0.89 (m) | 1.27 (overlap) | 1.29 (overlap) | 0.94 (d, 10.8) | 1.29 (overlap) | 1.30 (m) | 1.39 (d, 11.6) |
| 6 | 4.01 (td, 11.6, 4.0) | 5.23 (td, 11.2, 3.3) | 5.23 (td, 11.0, 3.2) | 3.91 (td, 10.8, 3.5) | 5.23 (td, 10.8, 3.5) | 5.22 (td, 11.2, 3.5) | 5.26 (td, 11.2, 3.7) |
| 7β | 2.18 (dd 11.6, 3.6) | 2.13 (dd, 11.6, 3.2) | 2.11 (dd, 12.0, 3.6) | 2.11 (dd, 12.0, 3.6) | 2.10 (dd, 12.0, 3.6) | 2.08 (overlap) | 2.07 (dd, 11.8, 3.4) |
| 7α | 1.00 (m) | 1.07 (overlap) | 1.06 (overlap) | 1.02 (overlap) | 1.09 (m) | 1.08 (overlap) | 1.19 (m) |
| 8 | - | - | - | - | - | - | - |
| 9 | 0.89 (m) | 1.40 (d, 11.2) | 1.38 (d, 11.2) | 1.31 (d, 11.2) | 1.38 (d, 10.8) | 1.36 (m) | 1.46 (d, 11.2) |
| 10 | - | - | - | - | - | - | - |
| 11 | 1.60 (m) 1.42 (m) | 5.49 (td, 11.2, 3.0) | 5.42 (td, 11.2, 2.8) | 5.40 (td, 10.8, 3.3) | 5.47 (td, 10.8, 3.6) | 5.42 (td, 11.0, 3.5) | 5.51 (td, 11.0, 3.6) |
| 12β | 2.09 (m) | 2.33 (dd, 12.4, 3.0) | 2.30 (dd, 12.0, 3.6) | 2.24 (dd, 11.6, 3.2) | 2.01 (m) | 1.98 (dd, 11.6, 3.6) | 2.65 (m) |
| 12α | 1.55 (m) | 1.75 (t, 11.2) | 1.69 (m) | 1.66 (t, 11.4) | 1.52 (m) | 1.52 (m) | 1.48 (m) |
| 13 | - | - | - | - | - | - | - |
| 14 | 1.22 (m) | 1.26 (dd, 11.6, 5.6) | 1.28 (overlap) | 1.22 (d, 12.0) | 1.25 (overlap) | 1.25 (overlap) | 1.32 (m) |
| 15 | 1.77 (m) 1.60 (m) | 1.75 (m) 1.60 (m) | 1.70 (m) 1.59 (m) | 1.81 (m) 1.56 (dd, 12.4, 6.4) | 1.88 (m) 1.53 (m) | 1.87 (m) 1.50 (m) | 1.94 (m) 1.86 (dd, 13.2, 6.4) |
| 16 | 2.77 (dd, 16.4, 6.4) 2.43 (dddd, 16.4, 12.4, 7.2, 1.6) | 2.76 (dd, 16.4, 5.6) 2.43 (m) | 2.76 (dd, 16.6, 6.2) 2.42 (m) | 2.72 (dd, 16.4, 6.0) 2.39 (m) | 2.42 (dd, 17.4, 5.0) 2.13 (m) | 2.41 (dd, 17.6, 4.8) 1.92 (m) | 2.53 (d, 5.2)/2.48 (d, 5.2) 2.34 (m) |
| 17 | - | - | - | - | - | - | - |
| 18 | - | - | - | - | - | - | - |
| 19 | 1.02 (s) | 0.87 (s) | 0.87 (s) | 0.98 (s) | 0.87 (s) | 0.87 (s) | 0.88 (s) |
| 20 | 1.15 (s) | 1.05 (s) | 1.05 (s) | 1.10 (s) | 1.06 (s) | 1.06 (s) | 1.09 (s) |
| 21 | 0.98 (s) | 1.15 (s) | 1.14 (s) | 1.04 (s) | 1.18 (s) | 1.15 (s) | 1.17 (s) |
| 22 | 0.90 (s) | 1.09 (s) | 1.07 (s) | 0.96 (s) | 1.08 (s) | 1.07 (s) | 1.09 (s) |
| 23 | 1.19 (s) | 1.31 (s) | 1.29 (s) | 1.27 (s) | 1.29 (s) | 1.25 (s) | 1.29 (s) |
| 24 | 7.04 (d, 1.2) | 7.04 (s) | 7.02 (d, 1.6) | 6.98 (s) | - | - | - |
| 25 | 7.07 (d, 1.2) | 7.07 (s) | 7.06 (d, 1.2) | 7.00 (s) | 4.73 (dt, 16.8, 2.8) 4.59 (ddd, 17.0, 3.8, 1.6) | 4.72 (d, 17.2) 4.62 (d, 16.8) | 5.84 (s) 5.82 (s) |
| 1′ | - | - | - | - | - | - | |
| 2′ | 3.43 (s) | 3.75 (d, 16.4) 3.49 (d, 16.0) | 5.69 (s) | 5.73 (d, 1.2) | 3.74 (d, 16.0) 3.53 (d, 16.0) | 5.75 (s) | |
| 3′ | - | - | - | - | - | - | |
| 4′ | - | 5.90 (s) | 3.10 (s) | 3.21 (s) | 5.91 (s) | 3.19 (s) | |
| 5′ | - | 2.02 (s) | - | - | 2.02 (s) | - | |
| 6′ | - | - | 2.23 (s) | 2.30 (d, 1.2) | - | 2.26 (d, 0.8) | |
| 1″ | - | - | - | - | - | - | |
| 2″ | 2.06 (s) | 2.05 (s) | - | 2.07 (s) | 2.06 (s) | 2.05 (s) |
| No. | 1 a | 2 a | 3 a | 4 b | 4 c | 5 a | 6 a | 7 d |
|---|---|---|---|---|---|---|---|---|
| 1 | 40.1 | 41.7 | 41.5 | 41.4 | 42.2 | 41.4 | 41.4 | 42.5 |
| 2 | 18.1 | 18.8 | 18.8 | 18.9 | 19.1 | 18.8 | 18.7 | 19.6 |
| 3 | 43.7 | 41.9 | 41.9 | 42.0 | 42.6 | 42.1 | 41.9 | 43.0 |
| 4 | 33.9 | 33.5 | 33.5 | 33.7 | 34.3 | 33.5 | 33.5 | 34.3 |
| 5 | 61.9 | 56.8 | 57.0 | 59.4 | 59.5 | 56.8 | 56.9 | 57.8 |
| 6 | 68.7 | 70.6 | 70.7 | 67.6 | 67.2 | 70.4 | 70.3 | 71.8 |
| 7 | 53.7 | 48.4 | 48.5 | 52.7 | 53.4 | 48.2 | 48.2 | 49.1 |
| 8 | 39.1 | 40.9 | 40.8 | 40.7 | 41.4 | 40.6 | 40.6 | 41.5 |
| 9 | 60.9 | 62.9 | 63.0 | 62.9 | 63.6 | 63.0 | 62.8 | 64.0/63.9 |
| 10 | 39.7 | 41.4 | 41.9 | 41.7 | 42.4 | 41.8 | 41.8 | 42.9 |
| 11 | 18.3 | 72.3 | 70.9 | 69.3 | 69.9 | 68.5 | 70.1 | 69.6 |
| 12 | 41.3 | 47.9 | 47.9 | 48.3 | 49.1 | 44.8 | 44.3 | 45.4/44.7 |
| 13 | 34.3 | 35.1 | 35.1 | 34.9 | 35.8 | 37.5 | 37.5 | 36.7/36.6 |
| 14 | 56.9 | 56.2 | 56.3 | 56.2 | 56.9 | 55.7 | 55.6 | 57.2 |
| 15 | 18.7 | 18.2 | 18.2 | 18.0 | 18.7 | 17.2 | 17.2 | 18.1/17.9 |
| 16 | 20.9 | 20.5 | 20.6 | 20.4 | 21.0 | 21.6 | 21.6 | 25.1/24.8 |
| 17 | 119.9 | 119.3 | 119.4 | 119.3, 119.1 | 118.3 | 123.9 | 123.8 | 137.9/137.8 |
| 18 | 137.6 | 136.3 | 136.5 | 136.6, 136.4 | 137.7 | 169.1 | 169.1 | 161.3/161.0 |
| 19 | 22.2 | 23.1 | 23.1 | 23.0 | 23.4 | 23.1 | 23.0 | 23.3 |
| 20 | 36.7 | 35.7 | 35.8 | 36.1 | 36.3 | 35.8 | 35.8 | 36.3 |
| 21 | 19.1 | 19.6 | 19.6 | 19.8 | 20.2 | 19.8 | 19.8 | 20.1 |
| 22 | 17.9 | 19.4 | 19.2 | 19.4 | 20.0 | 19.3 | 19.2 | 19.7 |
| 23 | 26.4 | 27.0 | 27.0 | 26.9 | 27.2 | 22.9 | 22.8 | 22.7 |
| 24 | 136.9 | 137.2 | 137.0 | 137.0, 136.9 | 138.0 | 174.1 | 174.2 | 171.8/171.4 |
| 25 | 135.2 | 135.1 | 135.2 | 135.0, 134.9 | 136.1 | 68.2 | 68.2 | 98.8/98.7 |
| 1′ | - | 166.6 | 169.3 | 165.5 | 166.0 | 165.1 | 169.3 | 166.8 |
| 2′ | - | 41.4 | 39.8 | 120.1 | 120.6 | 120.2 | 39.8 | 120.4 |
| 3′ | - | 170.0 | 154.0 | 152.0 | 153.1 | 151.9 | 153.8 | 154.6 |
| 4′ | - | - | 118.8 | 45.9 | 45.8 | 45.3 | 118.7 | 46.6 |
| 5′ | - | - | 170.9 | 172.6 | 171.4 | 173.0 | 169.9 | 173.8 |
| 6′ | - | - | 26.6 | 19.1 | 19.6 | 19.3 | 26.7 | 19.3 |
| 1″ | - | 170.6 | 170.5 | - | - | 170.5 | 170.5 | 172.2 |
| 2″ | - | 22.1 | 22.2 | - | - | 22.2 | 22.2 | 22.0 |
| Compound | A549 | HT29 | HeLa | HCT-116 | Vero Cells |
|---|---|---|---|---|---|
| 1 | 24.28 ± 7.56 | 22.71 ± 0.16 | 40.81 ± 0.73 | 20.28 ± 3.13 | >100 |
| 2 | 85.03 ± 10.17 | 33.02 ± 1.63 | 73.23 ± 5.37 | 14.33 ± 0.56 | >100 |
| 3 | 24.99 ± 1.59 | 20.75 ± 0.63 | 21.14 ± 1.66 | 21.41 ± 1.14 | 70.21 ± 8.23 |
| 4 | 22.95 ± 1.99 | 19.62 ± 2.32 | 23.72 ± 0.51 | 13.41 ± 0.39 | >100 |
| 5 | 99.14 ± 0.36 | 54.46 ± 5.93 | 72.92 ± 5.23 | 32.70 ± 2.37 | >100 |
| 6 | >100 | 78.25 ± 4.28 | 45.94 ± 2.77 | 16.53 ± 2.79 | >100 |
| 8 | 27.51 ± 0.32 | 20.39 ± 2.07 | 26.55 ± 2.58 | 24.41 ± 2.06 | >100 |
| cis-platin | 16.86 ± 0.03 | 5.30 ± 0.23 | 6.43 ± 0.13 | 4.93 ± 0.77 | >100 |
| Compound 1,2 | Staphylococcus aureus ATCC 25923 | Bacillus cereus ATCC 11778 | Bacillus subtilis ATCC 6633 | Methicillin Resistant Staphylococcus aureus DMST 20654 |
|---|---|---|---|---|
| 1 | 128 | inactive | inactive | inactive |
| 2 | 128 | 32 | 32 | 128 |
| 3 | inactive | 32 | 16 | inactive |
| 4 | 128 | inactive | inactive | inactive |
| 5 | 16 | 128 | 128 | inactive |
| 6 | 64 | 128 | inactive | inactive |
| 7 | inactive | 64 | 64 | inactive |
| 8 | 16 | 16 | 8 | 128 |
| Kanamycin | 2 | 16 | 8 | 4 |
| Chloramphenicol | 8 | 4 | 4 | 8 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wisetsai, A.; Lekphrom, R.; Bua-art, S.; Suebrasri, T.; Boonlue, S.; Tontapha, S.; Amornkitbamrung, V.; Senawong, T.; Schevenels, F.T. Scalarane Sesterterpenoids with Antibacterial and Anti-Proliferative Activities from the Mushroom Neonothopanus nambi. Molecules 2021, 26, 7667. https://doi.org/10.3390/molecules26247667
Wisetsai A, Lekphrom R, Bua-art S, Suebrasri T, Boonlue S, Tontapha S, Amornkitbamrung V, Senawong T, Schevenels FT. Scalarane Sesterterpenoids with Antibacterial and Anti-Proliferative Activities from the Mushroom Neonothopanus nambi. Molecules. 2021; 26(24):7667. https://doi.org/10.3390/molecules26247667
Chicago/Turabian StyleWisetsai, Awat, Ratsami Lekphrom, Sureeporn Bua-art, Thanapat Suebrasri, Sophon Boonlue, Sarawut Tontapha, Vittaya Amornkitbamrung, Thanaset Senawong, and Florian T. Schevenels. 2021. "Scalarane Sesterterpenoids with Antibacterial and Anti-Proliferative Activities from the Mushroom Neonothopanus nambi" Molecules 26, no. 24: 7667. https://doi.org/10.3390/molecules26247667
APA StyleWisetsai, A., Lekphrom, R., Bua-art, S., Suebrasri, T., Boonlue, S., Tontapha, S., Amornkitbamrung, V., Senawong, T., & Schevenels, F. T. (2021). Scalarane Sesterterpenoids with Antibacterial and Anti-Proliferative Activities from the Mushroom Neonothopanus nambi. Molecules, 26(24), 7667. https://doi.org/10.3390/molecules26247667