Phage-Display Based Discovery and Characterization of Peptide Ligands against WDR5
Abstract
:1. Introduction
2. Results and Discussion
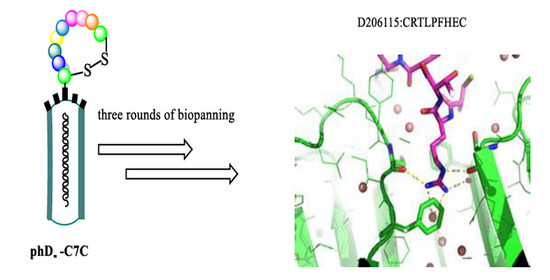
2.1. Affinity Selection of Cyclic Peptides Specific to WDR5
2.2. Measurement of Binding Affinity of Synthetic Peptides
2.3. Binding Interactions from the Crystal Structure
3. Materials and Methods
3.1. Bacterial Stains, Phage Library
3.2. Purified Protein for Biopanning and Crystallization
3.3. Biopanning of Phage-Displayed Cyclic Peptide
3.4. Sanger Sequencing
- Primer for PCR reaction:
- Forward C7C-1: 5′-GTCGGCGCAACTATCGGTATC-3′
- Reverse C7C-R1: 5′-GCCCTCATAGTTAGCGTAACG-3′
3.5. Synthetic Peptides and Fluorescence Polarization (FP) Binding Assays
3.6. Crystallization and Cocrystal Structure Determination
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Jain, B.P.; Pandey, S. WD40 repeat proteins: Signalling scaffold with diverse functions. Protein J. 2018, 37, 391–406. [Google Scholar] [CrossRef] [PubMed]
- Song, R.; Wang, Z.D.; Schapira, M. Disease Association and Druggability of WD40 Repeat Proteins. J. Proteome Res. 2017, 16, 3766–3773. [Google Scholar] [CrossRef] [PubMed]
- Migliori, V.; Mapelli, M.; Guccione, E. On WD40 proteins: Propelling our knowledge of transcriptional control? Epigenetics 2012, 7, 815–822. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guarnaccia, A.D.; Tansey, W.P. Moonlighting with WDR5: A cellular multitasker. J. Clin. Med. 2018, 7, 21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schapira, M.; Tyers, M.; Torrent, M.; Arrowsmith, C.H. WD40 repeat domain proteins: A novel target class? Nat. Rev. Drug Discov. 2017, 16, 773–786. [Google Scholar] [CrossRef]
- Wysocka, J.; Swigut, T.; Milne, T.A.; Dou, Y.; Zhang, X.; Burlingame, A.L.; Roeder, R.G.; Brivanlou, A.H.; Allis, C.D. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell 2005, 121, 859–872. [Google Scholar] [CrossRef] [Green Version]
- Roguev, A.; Schaft, D.; Shevchenko, A.; Pijnappel, W.W.M.P.; Wilm, M.; Aasland, R.; Stewart, A.F. The Saccharomyces cerevisiae Set1 complex includes an Ash2 homologue and methylates histone 3 lysine 4. EMBO J. 2001, 20, 7137–7148. [Google Scholar] [CrossRef]
- Su, J.M.; Wang, F.; Cai, Y.; Jin, J.J. The functional analysis of histone acetyltransferase MOF in tumorigenesis. Int. J. Mol. Sci. 2016, 17, 99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dias, J.; Nguyen, N.V.; Georgiev, P.; Gaub, A.; Brettschneider, J.; Cusack, S.; Kadlec, J.; Akhtar, A. Structural analysis of the KANSL1/WDR5/KANSL2 complex reveals that WDR5 is required for efficient assembly and chromatin targeting of the NSL complex. Genes Dev. 2014, 28, 929–942. [Google Scholar] [CrossRef] [Green Version]
- Bode, D.; Yu, L.; Tate, P.; Pardo, M.; Choudhary, J. Characterization of two distinct nucleosome remodeling and deacetylase (NuRD) complex assemblies in embryonic stem cells. Mol. Cell Proteom. 2016, 15, 878–891. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ee, L.S.; McCannell, K.N.; Tang, Y.; Fernandes, N.; Hardy, W.R.; Green, M.R.; Chu, F.; Fazzio, T.G. An embryonic stem cell-specific NuRD complex functions through interaction with WDR5. Stem Cell Rep. 2017, 8, 1488–1496. [Google Scholar] [CrossRef] [Green Version]
- Thomas, L.R.; Foshage, A.M.; Weissmiller, A.M.; Tansey, W.P. The MYC-WDR5 nexus and cancer. Cancer Res. 2015, 75, 4012–4015. [Google Scholar] [CrossRef] [Green Version]
- Cao, F.; Townsend, E.C.; Karatas, H.; Xu, J.; Li, L.; Lee, S.; Liu, L.; Chen, Y.; Ouillette, P.; Zhu, J.; et al. Targeting MLL1 H3K4 methyltransferase activity in mixed-lineage leukemia. Mol. Cell 2014, 53, 247–261. [Google Scholar] [CrossRef] [Green Version]
- Thiel, A.T.; Blessington, P.; Zou, T.; Feather, D.; Wu, X.J.; Yan, J.Z.; Zhang, H.; Liu, Z.G.; Ernst, P.; Koretzky, G.A.; et al. MLL-AF9-induced leukemogenesis requires coexpression of the wild-type Mll allele. Cancer Cell. 2010, 17, 148–159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grebien, F.; Vedadi, M.; Getlik, M.; Giambruno, R.; Grover, A.; Avellino, R.; Skucha, A.; Vittori, S.; Kuznetsova, E.; Smil, D.; et al. Pharmacological targeting of the Wdr5-MLL interaction in C/EBPα N-terminal leukemia. Nat. Chem. Biol. 2015, 11, 571–578. [Google Scholar] [CrossRef] [PubMed]
- Thomas, L.R.; Wang, Q.G.; Grieb, B.C.; Phan, J.; Foshage, A.M.; Sun, Q.; Olejniczak, E.T.; Clark, T.; Dey, S.; Lorey, S.; et al. Interaction with WDR5 promotes target gene recognition and tumorigenesis by MYC. Mol. Cell 2015, 58, 440–452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, F.; Jeon, K.O.; Salovich, J.M.; Macdonald, J.D.; Alvarado, J.; Gogliotti, R.D.; Phan, J.; Olejniczak, E.T.; Sun, Q.; Wang, S.; et al. Discovery of potent 2-aryl-6,7-dihydro-5 H-pyrrolo[1,2- a]imidazoles as WDR5-WIN-site inhibitors using fragment-based methods and structure-based design. J. Med. Chem. 2018, 61, 5623–5642. [Google Scholar] [CrossRef]
- Tian, J.; Teuscher, K.B.; Aho, E.R.; Alvarado, J.R.; Mills, J.J.; Meyers, K.M.; Gogliotti, R.D.; Han, C.; Macdonald, J.D.; Sai, J.; et al. Discovery and structure-based optimization of potent and selective WD repeat domain 5 (WDR5) inhibitors containing a dihydroisoquinolinone bicyclic core. J. Med. Chem. 2020, 63, 656–675. [Google Scholar] [CrossRef]
- Bratkovič, T. Progress in phage display: Evolution of the technique and its applications. Cell Mol. Life Sci. 2010, 67, 749–767. [Google Scholar] [CrossRef]
- Hamzeh-Mivehroud, M.; Alizadeh, A.A.; Morris, M.B.; Church, W.B.; Dastmalchi, S. Phage display as a technology delivering on the promise of peptide drug discovery. Drug Discov. Today 2013, 18, 1144–1157. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.T.; Man, Z.T.; Peng, C.L.; Wang, G.Z.; Sun, S. A specific affinity cyclic peptide enhances the adhesion, expansion and proliferation of rat bone mesenchymal stem cells on β-tricalcium phosphate scaffolds. Mol. Med. Rep. 2019, 20, 1157–1166. [Google Scholar] [CrossRef] [Green Version]
- Bailey, T.; Elkan, C. Fitting a mixture model by expectation maximization to discover motifs in bipolymers. In Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology, Stanford, CA, USA, 14–17 August 1994; AAAI Press: Menlo Park, CA, USA; pp. 28–36. Available online: https://www.researchgate.net/publication/303140247_Fitting_a_mixture_model_by_expectation_maximization_to_discover_motifs_in_bipolymers (accessed on 20 February 2021).
- Luck, K.; Travé, G. Phage display can select over-hydrophobic sequences that may impair prediction of natural domain–peptide interactions. Bioinformatics 2011, 27, 899–902. [Google Scholar] [CrossRef] [Green Version]
- Arachchige, D.; Margaret Harris, M.; Coon, Z.; Carlsen, J.; Holub, J.M. Role of single disulfide linkages in the folding and activity of scyllatoxin-based BH3 domain mimetics. J. Pept. Sci. 2017, 23, 367–373. [Google Scholar] [CrossRef]
- Macdonald, J.D.; Chacón Simon, S.; Han, C.; Wang, F.; Shaw, J.G.; Howes, J.E.; Sai, J.; Yuh, J.P.; Camper, D.; Alicie, B.M.; et al. Discovery and optimization of salicylic acid-derived sulfonamide inhibitors of the WD repeat-containing protein 5–MYC protein–protein interaction. J. Med. Chem. 2019, 62, 11232–11259. [Google Scholar] [CrossRef]
- Rentero Rebollo, I.; Heinis, C. Phage selection of bicyclic peptides. Methods 2013, 60, 46–54. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.L.; Sexton, D.J.; Skogerson, K.; Devlin, M. Novel peptide inhibitors of angiotensin-converting enzyme 2. J. Biol. Chem. 2003, 278, 15532–15540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, D.D.; Chen, W.L.; Wang, Z.H.; Xie, Y.; Xu, X.L.; Jiang, Z.-Y.; Zhang, X.; Guo, X.-K. High-affinity small molecular blockers of mixed lineage leukemia 1 (MLL1)-WDR5 interaction inhibit MLL1 complex H3K4 methyltransferase activity. Eur. J. Med. Chem. 2016, 124, 480–489. [Google Scholar] [CrossRef] [PubMed]
- Getlik, M.; Smil, D.; Zepeda-Velázquez, C.; Bolshan, Y.; Poda, G.; Wu, H.; Dong, A.; Kuznetsova, E.; Marcellus, R.; Senisterra, G.; et al. Structure-based optimization of a small molecule antagonist of the interaction between WD repeat-containing protein 5 (WDR5) and mixed-lineage leukemia 1 (MLL1). J. Med. Chem. 2016, 59, 2478–2496. [Google Scholar] [CrossRef]
- Ran, X.; Zhao, Y.; Liu, L.; Bai, L.; Yang, C.-Y.; Zhou, B.; Meagher, J.L.; Chinnaswamy, K.; Stuckey, J.A.; Wang, S. Structure-based design of γ-carboline analogues as potent and specific BET bromodomain inhibitors. J. Med. Chem. 2015, 58, 4927–4939. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lv, K.K.; Chen, W.C.; Chen, D.Q.; Mou, J.; Zhang, H.J.; Fan, T.T.; Li, Y.L.; Cao, D.Y.; Wang, X.; Chen, L.; et al. Rational design and evaluation of 6-(Pyrimidin-2-ylamino)-3,4-dihydroquinoxalin-2(1H)-ones as polypharmacological inhibitors of BET and kinases. J. Med. Chem. 2020, 63, 9787–9802. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.S.; Zhang, K.H.; Cui, Y.; Wang, Z.J.; Pan, Q.Y.; Liu, K.; Sun, B.; Zhou, H.; Li, M.J.; Xu, Q.; et al. Upgrade of macromolecular crystallography beamline BL17U1 at SSRF. Nucl. Sci. Tech. 2018, 29, 68. [Google Scholar] [CrossRef]
- Otwinowski, Z.; Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol. 1997, 276, 307–326. [Google Scholar] [CrossRef]
- Murshudov, G.N.; Vagin, A.A.; Dodson, E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997, 53, 240–255. [Google Scholar] [CrossRef] [PubMed]
- Adams, P.D.; Grosse-Kunstleve, R.W.; Hung, L.W.; Ioerger, T.R.; McCoy, A.J.; Moriarty, N.W.; Read, R.J.; Sacchettini, J.C.; Sauter, N.K.; Terwilliger, T.C. PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 2002, 58, 1948–1954. [Google Scholar] [CrossRef] [Green Version]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 486–501. [Google Scholar] [CrossRef] [PubMed] [Green Version]




| Input Titer (PFU) | Output Titer (PFU) | Recovery Efficiency | Fold Increase | |
|---|---|---|---|---|
| Round 1 | 1.0 × 1011 | 1.2 × 107 | 1.2 × 10−4 | 1 |
| Round 2 | 2.0 × 1011 | 7.5 × 107 | 3.8 × 10−4 | 3.1 |
| Round 3 | 2.0 × 1011 | 9.5 × 108 | 4.8 × 10−3 | 39.6 |
| Round 4 | 2.0 × 1011 | 2.3 × 109 | 1.2 × 10−2 | 95.8 |
| Round 5 | 2.0 × 1011 | 2.3 × 109 | 1.2 × 10−2 | 95.8 |
| Round 3 | Reads (Sanger) |
|---|---|
| CRTLPFHEC | 7/80 |
| CEKMVATHC | 4/80 |
| CRTLPWNQC | 3/80 |
| CRTIPFTHC | 2/80 |
| CRTMEYTSC | 2/80 |
| CRTLPYHLC | 2/80 |
| CRTQPYNQC | 1/80 |
| Partner | Motifs |
|---|---|
| MLL1 | GSARAEV |
| MLL2 | GCARSEP |
| MLL3 | GCARSEP |
| MLL4 | GAARAEV |
| SET1A | GSARSEG |
| SET1B | GCARSEG |
| H3 | ——ARTKQ |
| KANSL1 | VAARTRP |
| MBD3C | GAARCRV |
| KIF2A | GSARARP |
| Targeting WDR5 | EC50 (μM) | Ki (μM) |
|---|---|---|
| D206115-CRTLPFHEC | 5.9 ± 0.2 | 1.2 ± 0.04 |
| D206116-CEKMVATHC | N.D. | N.D. |
| D206179-CRTLPWNNC | 2.3 ± 0.6 | 0.5 ± 0.1 |
| D206180-CRTLPYGAC | 2.7 ± 0.2 | 0.5 ± 0.05 |
| D206181-CRTLPFGSC | 4.4 ± 0.4 | 0.9 ± 0.08 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cao, J.; Fan, T.; Li, Y.; Du, Z.; Chen, L.; Wang, Y.; Wang, X.; Shen, J.; Huang, X.; Xiong, B.; et al. Phage-Display Based Discovery and Characterization of Peptide Ligands against WDR5. Molecules 2021, 26, 1225. https://doi.org/10.3390/molecules26051225
Cao J, Fan T, Li Y, Du Z, Chen L, Wang Y, Wang X, Shen J, Huang X, Xiong B, et al. Phage-Display Based Discovery and Characterization of Peptide Ligands against WDR5. Molecules. 2021; 26(5):1225. https://doi.org/10.3390/molecules26051225
Chicago/Turabian StyleCao, Jiawen, Tiantian Fan, Yanlian Li, Zhiyan Du, Lin Chen, Ying Wang, Xin Wang, Jingkang Shen, Xun Huang, Bing Xiong, and et al. 2021. "Phage-Display Based Discovery and Characterization of Peptide Ligands against WDR5" Molecules 26, no. 5: 1225. https://doi.org/10.3390/molecules26051225
APA StyleCao, J., Fan, T., Li, Y., Du, Z., Chen, L., Wang, Y., Wang, X., Shen, J., Huang, X., Xiong, B., & Cao, D. (2021). Phage-Display Based Discovery and Characterization of Peptide Ligands against WDR5. Molecules, 26(5), 1225. https://doi.org/10.3390/molecules26051225