SARS-CoV-2 Main Protease Active Site Ligands in the Human Metabolome
Abstract
:1. Introduction
2. Results
2.1. The Docking Target
2.2. The Ligand Database
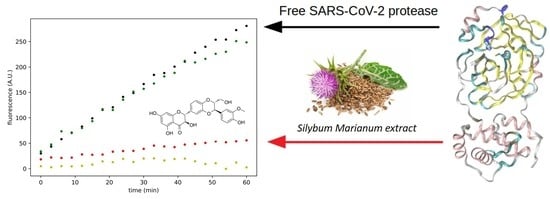
2.3. Effect of Silymarin on SARS-CoV-2 Mpro Activity
3. Discussion
4. Materials and Methods
4.1. In Silico Analyses
4.2. In Vitro Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, C.; Horby, P.W.; Hayden, F.G.; Gao, G.F. A novel coronavirus outbreak of global health concern. Lancet 2020, 395, 470–473. [Google Scholar] [CrossRef] [Green Version]
- Platto, S.; Wang, Y.; Zhou, J.; Carafoli, E. History of the covid-19 pandemic: Origin, explosion, worldwide spreading. Biochem. Biophys. Res. Commun. 2020, 538, 14–23. [Google Scholar] [CrossRef] [PubMed]
- Coronaviridae Study Group of the International Committee on Taxonomy of Viruses. The species severe acute respiratory syndrome-related coronavirus: Classifying 2019-ncov and naming it sars-cov-2. Nat. Microbiol. 2020, 5, 536–544. [Google Scholar] [CrossRef] [Green Version]
- Hadfield, J.; Megill, C.; Bell, S.M.; Huddleston, J.; Potter, B.; Callender, C.; Sagulenko, P.; Bedford, T.; Neher, R.A. Nextstrain: Real-time tracking of pathogen evolution. Bioinformatics 2018, 34, 4121–4123. [Google Scholar] [CrossRef]
- Andreano, E.; Piccini, G.; Licastro, D.; Casalino, L.; Johnson, N.V.; Paciello, I.; Dal Monego, S.; Pantano, E.; Manganaro, N.; Manenti, A.; et al. SARS-CoV-2 escape in vitro from a highly neutralizing COVID-19 convalescent plasma. bioRxiv 2020, 28, 424451. [Google Scholar] [CrossRef]
- Tegally, H.; Wilkinson, E.; Giovanetti, M.; Iranzadeh, A.; Fonseca, V.; Giandhari, J.; Doolabh, D.; Pillay, S.; San, E.J.; Msomi, N.; et al. Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. medRxiv 2020. [Google Scholar] [CrossRef]
- Caccuri, F.; Zani, A.; Messali, S.; Giovanetti, M.; Bugatti, A.; Campisi, G.; Filippini, F.; Scaltriti, E.; Ciccozzi, M.; Fiorentini, S.; et al. A persistently replicating SARS-CoV-2 variant derived from an asymptomatic individual. J. Transl. Med. 2020, 18, 362. [Google Scholar] [CrossRef]
- Plante, J.A.; Liu, Y.; Liu, J.; Xia, H.; Johnson, B.A.; Lokugamage, K.G.; Zhang, X.; Muruato, A.E.; Zou, J.; Fontes-Garfias, C.R.; et al. Spike mutation D614G alters SARS-CoV-2 fitness. Nature 2020. [Google Scholar] [CrossRef]
- Singh, J.; Ehtesham, N.Z.; Rahman, S.A.; Hasnain, S.E. Structure-function investigation of a new VUI-202012/01 SARS-CoV-2 variant. bioRxiv 2021. [Google Scholar] [CrossRef]
- Lai, M.M.; Cavanagh, D. The molecular biology of coronaviruses. Adv. Virus Res. 1997, 48, 1–100. [Google Scholar] [CrossRef]
- Masters, P.S. The molecular biology of coronaviruses. Adv.Virus Res. 2006, 66, 193–292. [Google Scholar] [CrossRef]
- Cui, J.; Li, F.; Shi, Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019, 17, 181–192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hilgenfeld, R. From SARS to MERS: Crystallographic studies on coronaviral proteases enable antiviral drug design. FEBS J. 2014, 281, 4085–4096. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anand, K.; Palm, G.J.; Mesters, J.R.; Siddell, S.G.; Ziebuhr, J.; Hilgenfeld, R. Structure of coronavirus main proteinase reveals combination of a chymotrypsin fold with an extra-helical domain. EMBO J. 2002, 21, 3213–3224. [Google Scholar] [CrossRef] [PubMed]
- Anand, K.; Ziebuhr, J.; Wadhwani, P.; Mesters, J.R.; Hilgenfeld, R. Coronavirus main proteinase (3CLpro) structure: Basis for design of anti-SARS drugs. Science 2003, 300, 1763–1767. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, F.; Chen, C.; Tan, W.; Yang, K.; Yang, H. Structure of main protease from human coronavirus NL63: Insights for wide spectrum anti-coronavirus drug design. Sci. Rep. 2016, 6, 22677. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Q.; Li, S.; Xue, F.; Zou, Y.; Chen, C.; Bartlam, M.; Rao, Z. Structure of the main protease from a global infectious human coronavirus, HCoV-HKU1. J. Virol. 2008, 82, 8647–8655. [Google Scholar] [CrossRef] [Green Version]
- Ratia, K.; Saikatendu, K.S.; Santarsiero, B.D.; Barretto, N.; Baker, S.C.; Stevens, R.C.; Mesecar, A.D. Severe acute respiratory syndrome coronavirus papain-like protease: Structure of a viral deubiquitinating enzyme. Proc. Natl. Acad. Sci. USA 2006, 103, 5717–5722. [Google Scholar] [CrossRef] [Green Version]
- Fu, L.; Ye, F.; Feng, Y.; Yu, F.; Wang, Q.; Wu, Y.; Zhao, C.; Sun, H.; Huang, B.; Niu, P.; et al. Both boceprevir and gc376 efficaciously inhibit sars-cov-2 by targeting its main protease. Nat. Commun. 2020, 11, 4417. [Google Scholar] [CrossRef]
- Vuong, W.; Khan, M.B.; Fischer, C.; Arutyunova, E.; Lamer, T.; Shields, J.; Saran, H.A.; McKay, R.T.; van Belkum, M.J.; Joyce, M.; et al. Feline coronavirus drug inhibits the main protease of sars-cov-2 and blocks virus replication. Nat. Commun. 2020, 11, 4282. [Google Scholar] [CrossRef]
- Watashi, K. Identifying and repurposing antiviral drugs against severe acute respiratory syndrome coronavirus 2 with in silico and in vitro approaches. Biochem. Biophys. Res. Commun. 2020, 538, 137–144. [Google Scholar] [CrossRef]
- Wishart, D.S.; Tzur, D.; Knox, C.; Eisner, R.; Guo, A.C.; Young, N.; Cheng, D.; Jewell, K.; Arndt, D.; Sawhney, S.; et al. HMDB: The Human Metabolome Database. Nucleic Acids Res. 2007, 35, D521–D526. [Google Scholar] [CrossRef]
- Viktorova, J.; Stranska-Zachariasova, M.; Fenclova, M.; Vitek, L.; Hajslova, J.; Kren, V.; Ruml, T. Complex evaluation of antioxidant capacity of milk thistle dietary supple-ments. Antioxid. 2019, 8, 317. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.-H.; Jassey, A.; Hsu, H.-Y.; Lin, L.-T. Antiviral Activities of Silymarin and Derivatives. Molecules 2019, 24, 1552. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bosch-Barrera, J.; Martin-Castillo, B.; Buxó, M.; Brunet, J.; Encinar, J.A.; Menendez, J.A. Silibinin and SARS-CoV-2: Dual Targeting of Host Cytokine Storm and Virus Replication Machinery for Clinical Management of COVID-19 Patients. J. Clin. Med. 2020, 9, 1770. [Google Scholar] [CrossRef] [PubMed]
- Jin, Z.; Du, X.; Xu, Y.; Deng, Y.; Liu, M.; Zhao, Y.; Zhang, B.; Li, X.; Zhang, L.; Peng, C.; et al. Structure of Mpro from sars-cov-2 and discovery of its inhibitors. Nature 2020, 582, 289–293. [Google Scholar] [CrossRef] [Green Version]
- Huang, C.; Wei, P.; Fan, K.; Liu, Y.; Lai, L. 3c-like proteinase from sars coronavirus catalyzes substrate hydrolysis by a general base mechanism. Biochem. 2004, 43, 4568–4574. [Google Scholar] [CrossRef] [PubMed]
- Palese, L.L. The structural landscape of sars-cov-2 main protease: Hints for inhibitor search. ChemRxiv 2020. [Google Scholar] [CrossRef]
- Fearon, D.; Owen, C.; Douangamath, A.; Lukacik, P.; Powell, A.; Strain-Damerell, C.; Resnick, E.; Krojer, T.; Gehrtz, P.; Wild, C.; et al. PanDDA Analysis Group Deposition of SARS-CoV-2 Main Protease Fragment Screen. Nat. Commun. 2020, 11, 5047. [Google Scholar] [CrossRef]
- Günaydın, Y.K.; Dündar, Z.D.; Çekmen, B.; Akıllı, N.B.; Köylü, R.; Cander, B. Intoxication due to Papaver rhoeas (Corn Poppy): Five Case Reports. Case Rep. Med. 2015, 2015. [Google Scholar] [CrossRef] [Green Version]
- Martorana, A.; Gentile, C.; Lauria, A. In Silico Insights into the SARS CoV-2 Main Protease Suggest NADH Endogenous Defences in the Control of the Pandemic Coronavirus Infection. Viruses 2020, 12, 805. [Google Scholar] [CrossRef] [PubMed]
- Verma, S.; Twilley, D.; Esmear, T.; Oosthuizen, C.B.; Reid, A.-M.; Nel, M.; Lall, N. Anti-SARS-CoV Natural Products With the Potential to Inhibit SARS-CoV-2 (COVID-19). Front. Pharmacol. 2020, 11, 561334. [Google Scholar] [CrossRef]
- Bhuiyan, F.R.; Howlader, S.; Raihan, T.; Hasan, M. Plants Metabolites: Possibility of Natural Therapeutics Against the COVID-19 Pandemic. Front. Med. 2020, 7, 444. [Google Scholar] [CrossRef]
- Chen, H.; Du, Q. Potential Natural Compounds for Preventing SARS-CoV-2 (2019-nCoV) Infection. Preprints 2020, 2020010358. [Google Scholar] [CrossRef]
- Jo, S.; Kim, H.; Kim, S.; Shin, D.H.; Kim, M.-S. Characteristics of flavonoids as potent MERS-CoV 3C-like protease inhibitors. Chem. Biol. Drug. Des. 2019, 94, 2023–2030. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Russo, M.; Moccia, S.; Spagnuolo, C.; Tedesco, I.; Russo, G.L. Roles of flavonoids against coronavirus infection. Chem. Biol. Interact. 2020, 328, 109211. [Google Scholar] [CrossRef]
- Pandit, M.; Latha, N. In silico studies reveal potential antiviral activity of phytochemicals from medicinal plants for the treatment of COVID-19 infection. Res Sq. 2020. [Google Scholar] [CrossRef] [Green Version]
- Debiaggi, M.; Tateo, F.; Pagani, L.; Luini, M.; Romero, E. Effects of propolis flavonoids on virus infectivity and replication. Microbiologica 1990, 13, 207–213. [Google Scholar]
- Abian, O.; Ortega-Alarcon, D.; Jimenez-Alesanco, A.; Ceballos-Laita, L.; Vega, S.; Reyburn, H.T.; Rizzuti, B.; Velazquez-Campoy, A. Structural stability of SARS-CoV-2 3CLpro and identification of quercetin as an inhibitor by experimental screening. Int. J. Biol. Macromol. 2020, 164, 1693–1703. [Google Scholar] [CrossRef]
- Valenzuela, A.; Aspillaga, M.; Vial, S.; Guerra, R. Selectivity of silymarin on the increase of the glutathione content in different tissues of the rat. Planta Med. 1989, 55, 420–422. [Google Scholar] [CrossRef]
- Fraschini, F.; Demartini, G.; Esposti, D. Pharmacology of Silymarin. Clin. Drug Investig. 2002, 22, 51–65. [Google Scholar] [CrossRef]
- Soleimani, V.; Delghandi, P.S.; Moallem, S.A.; Karimi, G. Safety and toxicity of silymarin, the major constituent of milk thistle extract: An updated review. Phytother Res. 2019, 33, 1627–1638. [Google Scholar] [CrossRef]
- Wishart, D.S.; Knox, C.; Guo, A.C.; Eisner, R.; Young, N.; Gautam, B.; Hau, D.D.; Psychogios, N.; Dong, E.; Bouatra, S.; et al. A knowledgebase for the human metabolome. Nucleic Acids Res. 2008, 37, D603–D610. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Jewison, T.; Guo, A.C.; Wilson, M.; Knox, C.; Liu, Y.; Djoumbou, Y.; Mandal, R.; Aziat, F.; Dong, E.; et al. HMDB 3.0—The Human Metabolome Database in 2013. Nucleic Acids Res. 2012, 41, D801–D807. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Feunang, Y.D.; Marcu, A.; Guo, A.C.; Liang, K.; Vàzquez-Fresno, R.; Sajed, T.; Johnson, D.; Li, C.; Karu, N.; et al. HMDB 4.0: The human metabolome database for 2018. Nucleic Acids Res. 2017, 46, D608–D617. [Google Scholar] [CrossRef] [PubMed]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug. Deliv. Rev. 2001, 46, 3–26. [Google Scholar] [CrossRef]
- Weininger, D. Smiles, a chemical language and information system. 1. introduction to methodology and encoding rules. J. Chem. Inf. Comput. Sci. 1988, 28, 31–36. [Google Scholar] [CrossRef]
- Sterling, T.; Irwin, J.J. Zinc 15-ligand discovery for everyone. J. Chem. Inf. Model. 2015, 55, 2324–2337. [Google Scholar] [CrossRef]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. Pubchem 2019 update: Improved access to chemical data. Nucleic Acids Res. 2019, 47, D1102–D1109. [Google Scholar] [CrossRef] [Green Version]
- Palese, L.L. Conformations of the HIV-1 protease: A crystal structure data set analysis. Biochim. Biophys. Acta 2017, 1865, 1416–1422. [Google Scholar] [CrossRef]
- Palese, L.L. Analysis of the conformations of the HIV-1 protease from a large crystallographic data set. Data. Brief. 2017, 15, 696–700. [Google Scholar] [CrossRef]
- Palese, L.L. A random version of principal component analysis in data clustering. Comput. Biol. Chem. 2018, 73, 57–64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [Green Version]
- Trott, O.; Olson, A.J. Autodock vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [Green Version]
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open babel: An open chemical toolbox. J. Cheminform. 2011, 3, 33. [Google Scholar] [CrossRef] [Green Version]
- Sanner, M.F. Python: A programming language for software integration and development. J. Mol. Graph. Model. 1999, 1, 57–61. [Google Scholar] [PubMed]
- Stoddard, S.V.; Stoddard, S.D.; Oelkers, B.K.; Fitts, K.; Whalum, K.; Whalum, K.; Hemphill, A.D.; Manikonda, J.; Martinez, L.M.; Riley, E.G.; et al. Optimization Rules for SARS-CoV-2 Mpro Antivirals: Ensemble Docking and Exploration of the Coronavirus Protease Active Site. Viruses 2020, 12, 942. [Google Scholar] [CrossRef]
- Keretsu, S.; Bhujbal, S.P.; Cho, S.J. Rational approach toward COVID-19 main protease inhibitors via molecular docking, mo-lecular dynamics simulation and free energy calculation. Sci. Rep. 2020, 10, 17716. [Google Scholar] [CrossRef]
- Phillips, J.C.; Braun, R.; Wang, W.; Gumbart, J.; Tajkhorshid, E.; Villa, E.; Chipot, C.; Skeel, R.D.; Kalé, L.; Schulten, K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005, 16, 1781–1802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bossis, F.; De Grassi, A.; Palese, L.L.; Pierri, C.L. Prediction of high- and low-affinity quinol-analogue-binding sites in the aa3 and bo3 terminal oxidases from Bacillus subtilis and Escherichia coli1. Biochem. J. 2014, 2, 305–314. [Google Scholar] [CrossRef] [PubMed]
- Jo, S.; Kim, T.; Iyer, V.G.; Im, W. CHARMM-GUI: A Web-based Graphical User Interface for CHARMM. J. Comput. Chem. 2008, 29, 1859–1865. [Google Scholar] [CrossRef] [PubMed]
- Brooks, B.R.; Brooks III, C.L.; MacKerell, A.D., Jr.; Nilsson, L.; Petrella, R.J.; Roux, B.; Won, Y.; Archontis, G.; Bartels, C.; Boresch, S.; et al. The Biomolecular Simulation Program. J. Comput. Chem. 2009, 30, 1545–1614. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Cheng, X.; Swails, J.M.; Yeom, M.S.; Eastman, P.K.; Lemkul, J.A.; Wei, S.; Buckner, J.; Jeong, J.C.; Qi, Y.; et al. CHARMM-GUI Input Generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM Simulations using the CHARMM36 Additive Force Field. J. Chem. Theory Comput. 2016, 12, 405–413. [Google Scholar] [CrossRef]
- Isgrò, C.; Sardanelli, A.M.; Palese, L.L. Systematic Search for SARS-CoV-2 Main Protease Inhibitors for Drug Repurposing: Ethacrynic Acid as a Potential Drug. Viruses 2021, 13, 106. [Google Scholar] [CrossRef]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sardanelli, A.M.; Isgrò, C.; Palese, L.L. SARS-CoV-2 Main Protease Active Site Ligands in the Human Metabolome. Molecules 2021, 26, 1409. https://doi.org/10.3390/molecules26051409
Sardanelli AM, Isgrò C, Palese LL. SARS-CoV-2 Main Protease Active Site Ligands in the Human Metabolome. Molecules. 2021; 26(5):1409. https://doi.org/10.3390/molecules26051409
Chicago/Turabian StyleSardanelli, Anna Maria, Camilla Isgrò, and Luigi Leonardo Palese. 2021. "SARS-CoV-2 Main Protease Active Site Ligands in the Human Metabolome" Molecules 26, no. 5: 1409. https://doi.org/10.3390/molecules26051409
APA StyleSardanelli, A. M., Isgrò, C., & Palese, L. L. (2021). SARS-CoV-2 Main Protease Active Site Ligands in the Human Metabolome. Molecules, 26(5), 1409. https://doi.org/10.3390/molecules26051409