Improved Diazo-Transfer Reaction for DNA-Encoded Chemistry and Its Potential Application for Macrocyclic DEL-Libraries
Abstract
:1. Introduction
2. Results and Discussion
2.1. Optimization of the Diazo-Transfer Reaction on Two Model Systems
2.2. Determining the Scope for the Optimized Diazo Reaction Conditions
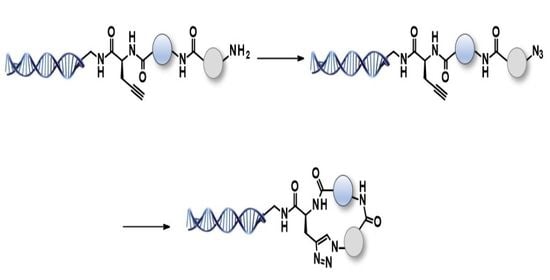
2.3. Off-DNA and on-DNA Macrocycle Formation
3. Materials and Methods
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Brenner, S.; Lerner, R.A. Encoded combinatorial chemistry. Proc. Natl. Acad. Sci. USA 1992, 89, 5381–5383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goodnow, R.A. A Handbook for DNA-Encoded Chemistry: Theory and Applications for Exploring Chemical Space and Drug Discovery; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2014. [Google Scholar]
- Goodnow, R.A., Jr.; Dumelin, C.E.; Keefe, A.D. DNA-encoded chemistry: Enabling the deeper sampling of chemical space. Nat. Rev. Drug Discov. 2017, 16, 131–147. [Google Scholar] [CrossRef]
- Castanon, J.; Roman, J.P.; Jessop, T.C.; de Blas, J.; Haro, R. Design and Development of a Technology Platform for DNA-Encoded Library Production and Affinity Selection. SLAS Discov. 2018, 23, 387–396. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lerner, R.A.; Brenner, S. DNA-Encoded Compound Libraries as Open Source: A Powerful Pathway to New Drugs. Angew. Chem. Int. Ed. 2017, 56, 1164–1165. [Google Scholar] [CrossRef] [Green Version]
- Harris, P.A.; Berger, S.B.; Jeong, J.U.; Nagilla, R.; Bandyopadhyay, D.; Campobasso, N.; Capriotti, C.A.; Cox, J.A.; Dare, L.; Dong, X.; et al. Discovery of a First-in-Class Receptor Interacting Protein 1 (RIP1) Kinase Specific Clinical Candidate (GSK2982772) for the Treatment of Inflammatory Diseases. J. Med. Chem. 2017, 60, 1247–1261. [Google Scholar] [CrossRef] [PubMed]
- Cuozzo, J.W.; Clark, M.A.; Keefe, A.D.; Kohlmann, A.; Mulvihill, M.; Ni, H.; Renzetti, L.M.; Resnicow, D.I.; Ruebsam, F.; Sigel, E.A.; et al. Novel Autotaxin Inhibitor for the Treatment of Idiopathic Pulmonary Fibrosis: A Clinical Candidate Discovered Using DNA-Encoded Chemistry. J. Med. Chem. 2020, 63, 7840–7856. [Google Scholar] [CrossRef] [PubMed]
- Satz, A.L.; Cai, J.; Chen, Y.; Goodnow, R.; Gruber, F.; Kowalczyk, A.; Petersen, A.; Naderi-Oboodi, G.; Orzechowski, L.; Strebel, Q. DNA Compatible Multistep Synthesis and Applications to DNA Encoded Libraries. Bioconjug. Chem. 2015, 26, 1623–1632. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Lundberg, H.; Asai, S.; Martín-Acosta, P.; Chen, J.S.; Brown, S.; Farrell, W.; Dushin, R.G.; O’Donnell, C.J.; Ratnayake, A.S.; et al. Kinetically guided radical-based synthesis of C(sp3)−C(sp3) linkages on DNA. Proc. Natl. Acad. Sci. USA 2018, 115, E6404–E6410. [Google Scholar] [CrossRef] [Green Version]
- Phelan, J.P.; Lang, S.B.; Sim, J.; Berritt, S.; Peat, A.J.; Billings, K.; Fan, L.; Molander, G.A. Open-Air Alkylation Reactions in Photoredox-Catalyzed DNA-Encoded Library Synthesis. J. Am. Chem. Soc. 2019, 141, 3723–3732. [Google Scholar] [CrossRef] [PubMed]
- Badir, S.O.; Sim, J.; Billings, K.; Csakai, A.; Zhang, X.; Dong, W.; Molander, G.A. Multifunctional Building Blocks Compatible with Photoredox-Mediated Alkylation for DNA-Encoded Library Synthesis. Org. Lett. 2020, 22, 1046–1051. [Google Scholar] [CrossRef]
- Kölmel, D.K.; Meng, J.; Tsai, M.-H.; Que, J.; Loach, R.P.; Knauber, T.; Wan, J.; Flanagan, M.E. On-DNA Decarboxylative Arylation: Merging Photoredox with Nickel Catalysis in Water. ACS Comb. Sci. 2019, 21, 588–597. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Roberts, S.E.; Franklin, G.J.; Davie, C.P. On-DNA Pd and Cu promoted C–N cross-coupling reactions. Med. Chem. Comm. 2017, 8, 1614–1617. [Google Scholar] [CrossRef]
- Ruff, Y.; Berst, F. Efficient copper-catalyzed amination of DNA-conjugated aryl iodides under mild aqueous conditions. Med. Chem. Commun. 2018, 9, 1188–1193. [Google Scholar] [CrossRef] [PubMed]
- De Pedro Beato, E.; Priego, J.; Gironda-Martínez, A.; González, F.; Benavides, J.; Blas, J.; Martín-Ortega, M.D.; Toledo, M.Á.; Ezquerra, J.; Torrado, A. Mild and Efficient Palladium-Mediated C–N Cross-Coupling Reaction between DNA-Conjugated Aryl Bromides and Aromatic Amines. ACS Comb. Sci. 2019, 21, 69–74. [Google Scholar] [CrossRef]
- Xuan Wang, X.; Sun, H.; Liu, J.; Zhong, W.; Zhang, M.; Zhou, H.; Dai, D.; Lu, X. Palladium-Promoted DNA-Compatible Heck Reaction. Org. Lett. 2019, 21, 719–723. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-C.; Faver, J.C.; Ku, A.F.; Miklossy, G.; Riehle, K.; Bohren, K.M.; Ucisik, M.N.; Matzuk, M.M.; Yu, Z.; Simmons, N. C–N Coupling of DNA-Conjugated (Hetero)aryl Bromides and Chlorides for DNA-Encoded Chemical Library Synthesis. Bioconjug. Chem. 2020, 31, 770–780. [Google Scholar] [CrossRef]
- Dillon, T.; Flood, D.T.; Asai, S.; Zhang, X.; Wang, J.; Yoon, L.; Adams, Z.C.; Dillingham, B.C.; Sanchez, B.B.; Vantourout, J.C.; et al. Expanding Reactivity in DNA-Encoded Library Synthesis via Reversible Binding of DNA to an Inert Quaternary Ammonium Support. J. Am. Chem. Soc. 2019, 141, 9998–10006. [Google Scholar]
- Ruff, Y.; Martinez, R.; Pellé, X.; Nimsgern, P.; Fille, P.; Ratnikov, M.; Berst, F. An Amphiphilic Polymer-Supported Strategy Enables Chemical Transformations under Anhydrous Conditions for DNA-Encoded Library Synthesis. ACS Comb. Sci. 2020, 22, 120–128. [Google Scholar] [CrossRef] [Green Version]
- Tanimoto, H.; Kakiuchi, K. Recent Applications and Developments of Organic Azides in Total Synthesis of Natural Products. Nat. Prod. Commun. 2013, 8, 1021–1034. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castro, V.; Blanco-Canosa, J.-B.; Rodriguez, H.; Albericio, F. Imidazole-1-sulfonyl Azide-Based Diazo-Transfer Reaction for the Preparation of Azido Solid Supports for Solid-Phase Synthesis. ACS Comb. Sci. 2013, 15, 331–334. [Google Scholar] [CrossRef]
- Remy Lartia, R.; Murat, P.; Dumy, P.; Defrancq, E. Versatile Introduction of Azido Moiety into Oligonucleotides through Diazo Transfer Reaction. Org. Lett. 2011, 13, 5672–5675. [Google Scholar] [CrossRef]
- Van Dongen, S.F.M.; Teeuwen, R.L.M.; Nallani, M.; van Berkel, S.S.; Cornelissen, J.J.L.M.; Nolte, R.J.M.; van Hest, J.C.M. Single-step azide introduction in proteins via an aqueous diazo transfer. Bioconjug. Chem. 2009, 20, 20–23. [Google Scholar]
- Gironda-Martínez, A.; Neri, D.; Samain, F.; Donckele, E.J. DNA-Compatible Diazo-Transfer Reaction in Aqueous Media Suitable for DNA-Encoded Chemical Library Synthesis. Org. Lett. 2019, 21, 9555–9558. [Google Scholar] [CrossRef]
- Fischer, N.; Goddard-Borger, E.D.; Greiner, R.; Klapötke, T.M.; Skelton, B.W.; Stierstorfer, J. Sensitivities of Some Imidazole-1-sulfonyl Azide Salts. J. Org. Chem. 2012, 77, 1760–1764. [Google Scholar] [CrossRef]
- Zhu, Z.; Shaginian, A.; Grady, L.C.; O’Keeffe, T.; Shi, X.E.; Davie, C.P.; Simpson, G.L.; Messer, J.A.; Evindar, G.; Bream, R.N.; et al. Design and Application of a DNA-Encoded Macrocyclic Peptide Library. ACS Chem. Biol. 2018, 13, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Clark, M.A.; Acharya, R.A.; Arico-Muendel, C.C.; Belyanskaya, S.L.; Benjamin, D.R.; Carlson, N.R.; Centrella, P.A.; Chiu, C.H.; Creaser, S.P.; Cuozzo, J.W.; et al. Design, synthesis and selection of DNA-encoded small-molecule libraries. Nat. Chem. Biol. 2009, 5, 647–654. [Google Scholar] [CrossRef] [PubMed]
- Litovchick, A.; Clark, M.A.; Keefe, A.D. Universal strategies for the DNA-encoding of libraries of small molecules using the chemical ligation of oligonucleotide tags. Artificial DNA: PNA XNA 2014, 5, e27896. [Google Scholar] [CrossRef] [Green Version]
- Andrew, R.; Bogdan, A.R.; Jerome, S.V.; Houk, K.N.; James, K. Strained Cyclophane Macrocycles: Impact of Progressive Ring Size Reduction on Synthesis and Structure. J. Am. Chem. Soc. 2012, 134, 2127–2138. [Google Scholar]



| Entry | ISA (Eq) | CuSO4 (Eq) | Buffer (Mol) | Temp. (°C) | Time (h) | Conversion a | |
|---|---|---|---|---|---|---|---|
| 1 to 3 (%) | 2 to 4 (%) | ||||||
| 1 | 50 | 10 | Borate (0.5) pH 9.4 | RT | 1 | 81 | 2 |
| 2 | 50 | 10 | RT | 16 | 100 | 39 | |
| 3 | 50 | 1 | RT | 16 | 61 | 49 | |
| 4 | 50 | 10 | NaHCO3 (0.2) | RT | 1 | 49 | 9 |
| 5 | 50 | 10 | RT | 16 | 98 | 55 | |
| 6 | 50 | 1 | RT | 16 | 100 | 48 | |
| 7 | 50 | 1 | 60 °C | 16 | 93 b | 50c | |
| 8 | 50 | 10 | K2CO3 (0.05) | RT | 16 | 98 | 96 |
| 9 | 50 | 1 | RT | 16 | 7 | 53 | |
| Entry | Substrate | Conversion b | Entry | Substrate | Conversion b | Entry | Substrate | Conversion b |
|---|---|---|---|---|---|---|---|---|
| 1 |  | 5% a 95% | 13 |  | 98% | 25 |  | 100% |
| 2 |  | 100% a 100% | 14 |  | 99% | 26 |  | 86% |
| 3 |  | 36% a 100% | 15 |  | 100% | 27 |  | 86% |
| 4 |  | 6% a 100% | 16 |  | 83% | 28 |  | 63% |
| 5 |  | 6% a 100% | 17 |  | 97% | 29 |  | 98% |
| 6 |  | 32% a 100% | 18 |  | 100% | 30 |  | 92% |
| 7 |  | 0% Control | 19 |  | 84% | 31 |  | 67% |
| 8 |  | 100% | 20 |  | 100% | 32 |  | 34% |
| 9 |  | 100% | 21 |  | 100% | 33 |  | 100% |
| 10 |  | 99% | 22 |  | 96% | 34 |  | 20% |
| 11 |  | 100% | 23 |  | 86% | 35 |  | 4% |
| 12 |  | 100% | 24 |  | 94% | 36 |  | 100% |
| Entry | 2nd Amino Acid  | 3rd Amino Acid  | Purity of 5 a (%) | Purity Azide 6 b (%) | Conversion | Ring Size |
|---|---|---|---|---|---|---|
| 6 to 7 (%) | Macrocycle 7 | |||||
| 1 | L-Phe | 3-Abz | 79 c | 74 c | 52 | n = 13 |
| 2 | L-Phe | 2-Abz | na | 88 d | 86 | n = 12 |
| 3 | L-Phe | β-Ala | 81 | 73 | 75 | n = 12 |
| 4 | L-Trp | β-Ala | 92 | 53 | 38 | n = 12 |
| 5 | L-Trp | Gly | 91 | 58 b | 0 | n = 11 |
| 6 | L-Trp | γ-Abu | 88 | 72 | 50 | n = 13 |
| 7 | β-Ala | β-Ala | 89 | 89 | 76 | n = 13 |
| 8 | β-Ala | γ-Abu | 91 | 91 | 80 | n = 14 |
| 9 | β-Ala | 3-Abz | 93 | 93 | 73 | n = 14 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ede, S.; Schenk, M.; Bierer, D.; Weinmann, H.; Graham, K. Improved Diazo-Transfer Reaction for DNA-Encoded Chemistry and Its Potential Application for Macrocyclic DEL-Libraries. Molecules 2021, 26, 1790. https://doi.org/10.3390/molecules26061790
Ede S, Schenk M, Bierer D, Weinmann H, Graham K. Improved Diazo-Transfer Reaction for DNA-Encoded Chemistry and Its Potential Application for Macrocyclic DEL-Libraries. Molecules. 2021; 26(6):1790. https://doi.org/10.3390/molecules26061790
Chicago/Turabian StyleEde, Selahattin, Mandy Schenk, Donald Bierer, Hilmar Weinmann, and Keith Graham. 2021. "Improved Diazo-Transfer Reaction for DNA-Encoded Chemistry and Its Potential Application for Macrocyclic DEL-Libraries" Molecules 26, no. 6: 1790. https://doi.org/10.3390/molecules26061790
APA StyleEde, S., Schenk, M., Bierer, D., Weinmann, H., & Graham, K. (2021). Improved Diazo-Transfer Reaction for DNA-Encoded Chemistry and Its Potential Application for Macrocyclic DEL-Libraries. Molecules, 26(6), 1790. https://doi.org/10.3390/molecules26061790