Antimicrobial Activity of Pyrazinamide Coordination Frameworks Synthesized by Mechanochemistry
Abstract
:1. Introduction
2. Results and Discussion
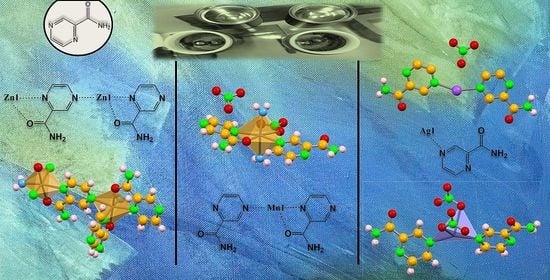
2.1. Structural Characterization
2.2. Shelf and Thermal Stability of Compounds 2, 3, and 5
2.3. Antimicrobial Activity of the Compounds
3. Experimental Details
3.1. Reagents
3.2. Synthesis of the Compounds
3.3. General Characterization
3.4. Single Crystal X-ray Diffraction Studies (SCXRD)
3.5. Antibacterial Activity Assays
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, Y.; Mitchison, D. The curious characteristics of pyrazinamide: A review. Int. J. Tuberc. Lung Dis. 2003, 7, 6–21. [Google Scholar] [PubMed]
- Somoskovi, A.; Wade, M.M.; Sun, Z.; Zhang, Y. Iron enhances the antituberculous activity of pyrazinamide. J. Antimicrob. Chemother. 2004, 53, 192–196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cavalcante, S.; Chakaya, J.M.; Egwaga, S.M.; Gie, R.; Gondrie, P.; Harries, A.D.; Hopewell, P.; Kumar, B.; Weezenbeck, K.L.-v.; Mase, S.; et al. Treatment of tuberculosis: Guidelines, 4th ed.; World Health Organization: Geneva, Switzerland, 2010. [Google Scholar]
- World Health Organization Model List of Essential Medicines, 21st List; World Health Organization: Geneva, Switzerland, 2019. Available online: https://apps.who.int/iris/bitstream/handle/10665/325771/WHO-MVP-EMP-IAU-2019.06-eng.pdf?ua=1 (accessed on 15 January 2021).
- A Mitchison, D.; Davies, G. The chemotherapy of tuberculosis: Past, present and future [State of the art]. Int. J. Tuberc. Lung Dis. 2012, 16, 724–732. [Google Scholar] [CrossRef]
- Eddaoudi, M.; Sava, D.F.; Eubank, J.F.; Adil, K.; Guillerm, V. Zeolite-like metal–organic frameworks (ZMOFs): Design, synthesis, and properties. Chem. Soc. Rev. 2015, 44, 228–249. [Google Scholar] [CrossRef] [Green Version]
- Guillerm, V.; Kim, D.; Eubank, J.F.; Luebke, R.; Liu, X.; Adil, K.; Lah, M.S.; Eddaoudi, M. A supermolecular building approach for the design and construction of metal–organic frameworks. Chem. Soc. Rev. 2014, 43, 6141–6172. [Google Scholar] [CrossRef] [Green Version]
- Eddaoudi, M.; Moler, D.B.; Li, H.; Chen, B.; Reineke, T.M.; O’Keeffe, M.; Yaghi, O.M. Modular Chemistry: Secondary Building Units as a Basis for the Design of Highly Porous and Robust Metal−Organic Carboxylate Frameworks. Accounts Chem. Res. 2001, 34, 319–330. [Google Scholar] [CrossRef]
- Miller, S.R.; Heurtaux, D.; Baati, T.; Horcajada, P.; Grenèche, J.-M.; Serre, C. Biodegradable therapeutic MOFs for the delivery of bioactive molecules. Chem. Commun. 2010, 46, 4526–4528. [Google Scholar] [CrossRef]
- Li, S.; Huo, F. Metal–organic framework composites: From fundamentals to applications. Nanoscale 2015, 7, 7482–7501. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Farha, O.K.; Roberts, J.; Scheidt, K.A.; Nguyen, S.T.; Hupp, J.T. Metal–organic framework materials as catalysts. Chem. Soc. Rev. 2009, 38, 1450–1459. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.-Y.; Qin, C.; Wang, X.-L.; Su, Z.-M. Metal-organic frameworks as potential drug delivery systems. Expert Opin. Drug Deliv. 2012, 10, 89–101. [Google Scholar] [CrossRef]
- Wang, S.; Wang, X. Multifunctional Metal-Organic Frameworks for Photocatalysis. Small 2015, 11, 3097–3112. [Google Scholar] [CrossRef]
- Wuttke, S.; Braig, S.; Preiß, T.; Zimpel, A.; Sicklinger, J.; Bellomo, C.; Rädler, J.O.; Vollmar, A.M.; Bein, T. MOF nanoparticles coated by lipid bilayers and their uptake by cancer cells. Chem. Commun. 2015, 51, 15752–15755. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.-G.; Dong, Z.-Y.; Cheng, H.; Wan, S.-S.; Chen, W.-H.; Zou, M.-Z.; Huo, J.-W.; Deng, H.-X.; Zhang, X.-Z. A multifunctional metal–organic framework based tumor targeting drug delivery system for cancer therapy. Nanoscale 2015, 7, 16061–16070. [Google Scholar] [CrossRef]
- Imaz, I.; Rubio-Martínez, M.; An, J.; Solé-Font, I.; Rosi, N.L.; Maspoch, D. Metal–biomolecule frameworks (MBioFs). Chem. Commun. 2011, 47, 7287–7302. [Google Scholar] [CrossRef]
- Horcajada, P.; Gref, R.; Baati, T.; Allan, P.K.; Maurin, G.; Couvreur, P.; Férey, G.; Morris, R.E.; Serre, C. Metal–Organic Frameworks in Biomedicine. Chem. Rev. 2011, 112, 1232–1268. [Google Scholar] [CrossRef]
- McKinlay, A.C.; Morris, R.E.; Horcajada, P.; Férey, G.; Gref, R.; Couvreur, P.; Serre, C. BioMOFs: Metal-Organic Frameworks for Biological and Medical Applications. Angew. Chem. Int. Ed. 2010, 49, 6260–6266. [Google Scholar] [CrossRef] [PubMed]
- Horcajada, P.; Serre, C.; Vallet-Regí, M.; Sebban, M.; Taulelle, F.; Férey, G. Metal–Organic Frameworks as Efficient Materials for Drug Delivery. Angew. Chem. Int. Ed. 2006, 45, 5974–5978. [Google Scholar] [CrossRef] [PubMed]
- Horcajada, P.; Serre, C.; Maurin, G.; Ramsahye, N.A.; Balas, F.; Vallet-Regí, M.; Sebban, M.; Taulelle, F.; Férey, G. Flexible Porous Metal-Organic Frameworks for a Controlled Drug Delivery. J. Am. Chem. Soc. 2008, 130, 6774–6780. [Google Scholar] [CrossRef] [PubMed]
- Babarao, R.; Jiang, J. Unraveling the Energetics and Dynamics of Ibuprofen in Mesoporous Metal−Organic Frameworks. J. Phys. Chem. C 2009, 113, 18287–18291. [Google Scholar] [CrossRef]
- An, J.; Geib, S.J.; Rosi, N.L. Cation-Triggered Drug Release from a Porous Zinc−Adeninate Metal−Organic Framework. J. Am. Chem. Soc. 2009, 131, 8376–8377. [Google Scholar] [CrossRef]
- Alves, P.C.; Rijo, P.; Bravo, C.; Antunes, A.M.M.; André, V. Bioactivity of Isostructural Hydrogen Bonding Frameworks Built from Pipemidic Acid Metal Complexes. Molecules 2020, 25, 2374. [Google Scholar] [CrossRef]
- Andre, V.; Da Silva, A.R.F.; Fernandes, A.; Frade, R.; Garcia, C.; Rijo, P.; Antunes, A.M.M.; Rocha, J.; Duarte, M.T. Mg- and Mn-MOFs Boost the Antibiotic Activity of Nalidixic Acid. ACS Appl. Bio Mater. 2019, 2, 2347–2354. [Google Scholar] [CrossRef]
- Quaresma, S.; André, V.; Antunes, A.M.M.; Vilela, S.M.F.; Amariei, G.; Arenas-Vivo, A.; Rosal, R.; Horcajada, P.; Duarte, M.T. Novel Antibacterial Azelaic Acid BioMOFs. Cryst. Growth Des. 2019, 20, 370–382. [Google Scholar] [CrossRef]
- André, V.; Galego, F.; Martins, M. Mechanochemical Assembly of Nalidixic Acid Bioinspired Metal–Organic Compounds and Complexes toward Improved Solubility. Cryst. Growth Des. 2018, 18, 2067–2081. [Google Scholar] [CrossRef]
- Amidon, G.L.; Lennernäs, H.; Shah, V.P.; Crison, J.R. A Theoretical Basis for a Biopharmaceutic Drug Classification: The Correlation of in Vitro Drug Product Dissolution and in Vivo Bioavailability. Pharm. Res. 1995, 12, 413–420. [Google Scholar] [CrossRef] [Green Version]
- Khadka, P.; Ro, J.; Kim, H.; Kim, I.; Kim, J.T.; Kim, H.; Cho, J.M.; Yun, G.; Lee, J. Pharmaceutical particle technologies: An approach to improve drug solubility, dissolution and bioavailability. Asian J. Pharm. Sci. 2014, 9, 304–316. [Google Scholar] [CrossRef] [Green Version]
- Hu, J.; Johnston, K.P.; Williams, R.O. Nanoparticle Engineering Processes for Enhancing the Dissolution Rates of Poorly Water Soluble Drugs. Drug Dev. Ind. Pharm. 2004, 30, 233–245. [Google Scholar] [CrossRef] [PubMed]
- Costa, P.; Lobo, J.M.S. Modeling and comparison of dissolution profiles. Eur. J. Pharm. Sci. 2001, 13, 123–133. [Google Scholar] [CrossRef]
- Salehi, S.; Moghaddam, S.M.M.; Tarin, M.; Saljooghi, A.S. Pharmaceutical Nickel (II) Chelation Properties of 3-Hydroxyflaven, Deferiprone and Maltol Metal Chelators: A Density Functional Study. Phys. Chem. Res. 2020, 8, 91–110. [Google Scholar] [CrossRef]
- Tran, Q.H.; Doan, T.T. A novel study on curcumin metal complexes: Solubility improvement, bioactivity, and trial burn wound treatment in rats. New J. Chem. 2020, 44, 13036–13045. [Google Scholar] [CrossRef]
- Abdolmaleki, S.; Ghadermazi, M.; Ashengroph, M.; Saffari, A.; Sabzkohi, S.M. Cobalt (II), zirconium(IV), calcium(II) complexes with dipicolinic acid and imidazole derivatives: X-ray studies, thermal analyses, evaluation as in vitro antibacterial and cytotoxic agents. Inorganica Chim. Acta 2018, 480, 70–82. [Google Scholar] [CrossRef]
- Allen, F.H. The Cambridge Structural Database: A quarter of a million crystal structures and rising. Acta Crystallogr. Sect. B Struct. Sci. 2002, 58, 380–388. [Google Scholar] [CrossRef]
- Tabrizi, L.; Chiniforoshan, H.; McArdle, P. A novel one-dimensional manganese(II) coordination polymer containing both dicyanamide and pyrazinamide ligands: Synthesis, spectroscopic investigations, X-ray studies and evaluation of biological activities. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2015, 139, 307–312. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.; Ahmed, M.; Hafiz, S.; Kamal, M.; Mumtaz, M.; Ayatollahi, S.A. Design, Synthesis and Antitubercular Evaluation of Novel Series of Pyrazinecarboxamide Metal Complexes. IJPR 2018, 17, 93–99. [Google Scholar]
- Stenger-Smith, J.; Kamariza, M.; Chakraborty, I.; Ouattara, R.; Bertozzi, C.R.; Mascharak, P.K. Enhanced Bactericidal Effects of Pyrazinamide Toward Mycobacterium smegmatis and Mycobacterium tuberculosis upon Conjugation to a {Au(I)-triphenylphosphine}+ Moiety. ACS Omega 2020, 5, 6826–6833. [Google Scholar] [CrossRef]
- Titi, H.M.; Do, J.-L.; Howarth, A.J.; Nagapudi, K.; Friščić, T. Simple, scalable mechanosynthesis of metal–organic frameworks using liquid-assisted resonant acoustic mixing (LA-RAM). Chem. Sci. 2020, 11, 7578–7584. [Google Scholar] [CrossRef] [Green Version]
- Friščić, T.; Mottillo, C.; Titi, H.M. Mechanochemistry for Synthesis. Angew. Chem. Int. Ed. 2020, 59, 1018–1029. [Google Scholar] [CrossRef] [PubMed]
- Hasa, D.; Rauber, G.S.; Voinovich, D.; Jones, W. Cocrystal Formation through Mechanochemistry: From Neat and Liquid-Assisted Grinding to Polymer-Assisted Grinding. Angew. Chem. Int. Ed. 2015, 54, 7371–7375. [Google Scholar] [CrossRef]
- James, S.L.; Adams, C.J.; Bolm, C.; Braga, D.; Collier, P.; Friščić, T.; Grepioni, F.; Harris, K.D.M.; Hyett, G.; Jones, W.; et al. Mechanochemistry: Opportunities for new and cleaner synthesis. Chem. Soc. Rev. 2011, 41, 413–447. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hasa, D.; Carlino, E.; Jones, W. Polymer-Assisted Grinding, a Versatile Method for Polymorph Control of Cocrystallization. Cryst. Growth Des. 2016, 16, 1772–1779. [Google Scholar] [CrossRef]
- Friščić, T. New opportunities for materials synthesis using mechanochemistry. J. Mater. Chem. 2010, 20, 7599–7605. [Google Scholar] [CrossRef]
- Friščić, T. Supramolecular concepts and new techniques in mechanochemistry: Cocrystals, cages, rotaxanes, open metal–organic frameworks. Chem. Soc. Rev. 2012, 41, 3493–3510. [Google Scholar] [CrossRef] [PubMed]
- Do, J.-L.; Friščić, T. Mechanochemistry: A Force of Synthesis. ACS Central Sci. 2017, 3, 13–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quaresma, S.; André, V.; Fernandes, A.; Duarte, M.T. Mechanochemistry – A green synthetic methodology leading to metallodrugs, metallopharmaceuticals and bio-inspired metal-organic frameworks. Inorganica Chim. Acta 2017, 455, 309–318. [Google Scholar] [CrossRef]
- Altaf, M.; Miller, C.H.; Bellows, D.S.; O’Toole, R. Evaluation of the Mycobacterium smegmatis and BCG models for the discovery of Mycobacterium tuberculosis inhibitors. Tuberc. 2010, 90, 333–337. [Google Scholar] [CrossRef]
- Etienne, G.; Laval, F.; Villeneuve, C.; Dinadayala, P.; Abouwarda, A.; Zerbib, D.; Galamba, A.; Daffé, M. The cell envelope structure and properties of Mycobacterium smegmatis mc2155: Is there a clue for the unique transformability of the strain? Microbiol. 2005, 151, 2075–2086. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shirvan, S.A.; Dezfuli, S.H. catena-Poly[[[aqua(pyrazine-2-carboxamide-κ2N1,O)zinc]-μ-pyrazine-2-carboxamide-κ3N1,O:N4] dinitrate]. Acta Crystallogr. Sect. E Struct. Rep. Online 2012, 68, m627–m628. [Google Scholar] [CrossRef] [Green Version]
- Yamada, H.; Yamaguchi, M.; Igarashi, Y.; Chikamatsu, K.; Aono, A.; Murase, Y.; Morishige, Y.; Takaki, A.; Chibana, H.; Mitarai, S. Mycolicibacterium smegmatis, Basonym Mycobacterium smegmatis, Expresses Morphological Phenotypes Much More Similar to Escherichia coli Than Mycobacterium tuberculosis in Quantitative Structome Analysis and CryoTEM Examination. Front. Microbiol. 2018, 9, 1992. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, L.M.; Fu-Liu, C.S. Is Mycobacterium tuberculosis a closer relative to Gram-positive or Gram–negative bacterial pathogens? Tuberculosis 2002, 82, 85–90. [Google Scholar] [CrossRef]
- Hett, E.C.; Rubin, E.J. Bacterial Growth and Cell Division: A Mycobacterial Perspective. Microbiol. Mol. Biol. Rev. 2008, 72, 126–156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Macrae, C.F.; Bruno, I.J.; Chisholm, J.A.; Edgington, P.R.; McCabe, P.; Pidcock, E.; Rodriguez-Monge, L.; Taylor, R.J.; Van De Streek, J.; Wood, P.A. Mercury CSD 2.0– new features for the visualization and investigation of crystal structures. J. Appl. Crystallogr. 2008, 41, 466–470. [Google Scholar] [CrossRef]
- Bruker AXS:SAINT+, release 6.22; Bruker Analytical Systems: Madison, WI, USA, 2005.
- Bruker AXS:SADABS; Bruker Analytical Systems: Madison, WI, USA, 2005.
- Sheldrick, G.M. SHELXT– Integrated space-group and crystal-structure determination. Acta Cryst. 2015, 71, 3–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sheldrick, G.M. Crystal structure refinement with SHELXL. Acta Cryst. 2015, 71, 3–8. [Google Scholar] [CrossRef]
- Farrugia, L.J. WinGX suite for small-molecule single-crystal crystallography. J. Appl. Crystallogr. 1999, 32, 837–838. [Google Scholar]
- Spek, A.L. Single-crystal structure validation with the programPLATON. J. Appl. Crystallogr. 2003, 36, 7–13. [Google Scholar] [CrossRef] [Green Version]
- Siopa, F.; Figueiredo, T.; Frade, R.F.M.; Neto, I.; Meirinhos, A.; Reis, C.P.; Sobral, R.G.; Afonso, C.A.M.; Rijo, P. Choline-Based Ionic Liquids: Improvement of Antimicrobial Activity. ChemistrySelect 2016, 1, 5909–5916. [Google Scholar] [CrossRef]
- CLSI. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically, 11th ed.; CLSI: Wayne, PA, USA, 2018. [Google Scholar]

















| Compound | Symmetry Operation | D–H⋯A | d(D-H) (Å) | d(H⋯A) (Å) | d(D⋯A) (Å) | (DHA) (°) |
|---|---|---|---|---|---|---|
| 1 | 1 − x,1 − y, 1 − z | N3–H7⋯O3W | 0.86 | 2.50 | 3.193(8) | 138 |
| 1 − x,1 − y, 1 − z | N3–H7⋯O5 | 0.86 | 2.42 | 3.163(8) | 144 | |
| x, y, z | N3–H8⋯O7 | 0.86 | 2.08 | 2.937(8) | 172 | |
| 2 − x, 1 − y, 1 − z | N6–H9⋯O5 | 0.86 | 2.41 | 3.231(8) | 161 | |
| 2 − x, ½ + y, ½ − z | N6–H10⋯O4 | 0.86 | 2.07 | 2.913(7) | 166 | |
| x, y, z | O3W–H11W⋯O4 | 0.75(6) | 2.06(7) | 2.781(8) | 162(9) | |
| 1 − x,1 − y, 1 − z | O3W–H12W⋯O8 | 0.76(7) | 2.05(7) | 2.810(8) | 177(7) | |
| 2 | 1 − x, 2 − y, − z | N3–H1N⋯O8 | 0.77(2) | 2.15(3) | 2.889(2) | 159(2) |
| 2 − x, 1 − y, 1 − z | O1w–H1w⋯O2 | 0.86(3) | 1.94(3) | 2.7773(18) | 166(2) | |
| x, 1 + y, z | N3–H2N⋯O4 | 0.89(3) | 2.54(2) | 3.368(2) | 155(2) | |
| x, 1 + y, z | N3–H2N⋯O5 | 0.89(3) | 2.41(3) | 3.228(2) | 153(2) | |
| x, y, z | O1w–H2w⋯O6 | 0.81(3) | 1.87(3) | 2.6723(19) | 169(3) | |
| 1 + x, y, 1 + z | N6–H3N⋯N2 | 0.81(3) | 2.23(2) | 3.008(2) | 163(2) | |
| 1 − x, 1 − y, 1 − z | O2w–H3w⋯O3 | 0.75(3) | 2.28(3) | 2.9714(18) | 155(3) | |
| 1 − x, 1 − y, 1 − z | O2w–H3w⋯O4 | 0.75(3) | 2.51(3) | 3.186(2) | 150(3) | |
| 2 − x, 2 − y, 1 − z | N6–H4N⋯O7 | 0.87(3) | 2.20(3) | 2.880(2) | 135(2) | |
| 1 − x, 1 − y, 1 − z | O2w–H4w⋯O2 | 0.89(3) | 1.84(3) | 2.7278(17) | 178(3) | |
| 3 | x, y, z | N3–H1N⋯N2 | 0.90(5) | 2.44(8) | 2.733(7) | 100(6) |
| 2 − x 1 − y, 1 − z | N3–H1N⋯N2 | 0.90(5) | 2.44(8) | 3.150(7) | 137(6) | |
| 1 − x, 1 − y, 2 − z | N3–H2N⋯O1 | 0.90(6) | 2.02(6) | 2.888(7) | 164(6) | |
| 4 | x, − y, 1 − z | N3–H1N⋯N2 | 0.81(8) | 2.48(8) | 3.118(8) | 136(7) |
| 2 − x, 1 − y, 1 − z | N3–H2N⋯O1 | 0.79(8) | 2.10(8) | 2.883(8) | 177(8) |
| Compounds | E. coli | S. aureus | M. smegmatis | |||
|---|---|---|---|---|---|---|
| MIC * (µg/mL) | MBC * (µg/mL) | MIC ** (µg/mL) | MBC * (µg/mL) | MIC ** (µg/mL) | MBC * (µg/mL) | |
| Pyrazinamide | >62.5 | 250 | >250 | 500 | >62.5 | 125 |
| (1) | >62.5 | 500 | >250 | 500 | >62.5 | 62.5 |
| Zn(NO3)2 | >62.5 | 500 | >250 | 500 | >62.5 | 62.5 |
| (2) | >62.5 | 500 | >250 | 500 | >62.5 | 125 |
| Mn(NO3)2 | >62.5 | 500 | >250 | 500 | >62.5 | 125 |
| (3) | 15.63 | 31.25 | 62.5 | 125 | 15.63 | 31.25 |
| (5) | 7.81 | 31.25 | 62.5 | 125 | 7.81 | 31.25 |
| AgNO3 | 7.81 | 31.25 | 62.5 | 125 | 7.81 | 7.81 |
| Positive control | <0.49 (NOR) | Nd | 0.98 (VAN) | Nd | <0.49 (VAN) | Nd |
| Negative control (DMSO) | 62.5 | Nd | 250 | Nd | 62.5 | Nd |
| Pyrazinamide (PYR) | Metal Salt (M) | Molar Ratio (PYR:M) | Time | Resulting Compound |
|---|---|---|---|---|
| 0.0907 g (0.74 mmol) | Zn(NO3)2⋅6H2O 0.1105 g (0.37 mmol) | 2:1 | 10 min | (1) |
| 0.0992 g (0.81 mmol) | Mn(NO3)2⋅4H2O 0.1036 g (0.41 mmol) | 2:1 | 15 min | (2) |
| 0.1183 g (0.96 mmol) | AgNO3 0.0829 g (0.49 mmol) | 2:1 | (3) | |
| 0.0840 g (0.68 mmol) | AgNO3 0.1171 g (0.69 mmol) | 1:1 | (5) |
| 2 | 3 | 4 | |
|---|---|---|---|
| Chemical formula | C10H10N6O2Mn⋅2H2O⋅2NO3 | C10H10N6O2Ag⋅NO3 | C10H10N6O2Ag⋅NO3 |
| Formula weight | 461.23 | 416.12 | 416.12 |
| Temperature (K) | 293 | 293 | 293 |
| Wavelength (Å) | 0.71073 | 0.71073 | 0.71073 |
| Crystal form, color | Block, colorless | Block, colorless | Plate, colorless |
| Crystal size (mm) | 0.17 × 0.08 × 0.03 | 0.14 × 0.10 × 0.10 | 0.12 × 0.12 × 0.10 |
| Crystal system | Triclinic | Monoclinic | Monoclinic |
| Space group | P | P21/m | P2/c |
| a (Å) | 6.9866(3) | 3.6132(11) | 3.5903(4) |
| b (Å) | 9.2273(4) | 27.735(8) | 6.7181(6) |
| c (Å) | 14.1882(5) | 6.6273(19) | 27.610(3) |
| α (°) | 90.584(3) | 90.00 | 90.00 |
| β (°) | 99.285(2) | 94.007(9) | 91.149(5) |
| γ (°) | 101.460(2) | 90.00 | 90.00 |
| V (Å3) | 883.85(6) | 662.5(3) | 665.81(11) |
| Z | 2 | 2 | 2 |
| d (mg.m−3) | 1.733 | 2.086 | 2.076 |
| µ (mm−1) | 0.820 | 1.564 | 1.556 |
| F(000) | 470 | 412 | 412 |
| θ range (°) | 2.630–30.976 | 3.081–26.510 | 2.952–26.563 |
| Reflections collected/unique | 15432/5513 | 4443/1381 | 5186/1374 |
| Rint | 0.0349 | 0.0470 | 0.0301 |
| GOF | 1.020 | 1.132 | 1.217 |
| Final R indices [I > 2σ(I)] | R1 = 0.0357, wR2 = 0.0892 | R1 = 0.0630, wR2 = 0.1316 | R1 = 0.0577, wR2 = 0.1256 |
| Indices all data | R1 = 0.0476, wR2 = 0.0938 | R1 = 0.0780, wR2 = 0.1372 | R1 = 0.0620, wR2 = 0.1273 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Quaresma, S.; Alves, P.C.; Rijo, P.; Duarte, M.T.; André, V. Antimicrobial Activity of Pyrazinamide Coordination Frameworks Synthesized by Mechanochemistry. Molecules 2021, 26, 1904. https://doi.org/10.3390/molecules26071904
Quaresma S, Alves PC, Rijo P, Duarte MT, André V. Antimicrobial Activity of Pyrazinamide Coordination Frameworks Synthesized by Mechanochemistry. Molecules. 2021; 26(7):1904. https://doi.org/10.3390/molecules26071904
Chicago/Turabian StyleQuaresma, Sílvia, Paula C. Alves, Patrícia Rijo, M. Teresa Duarte, and Vânia André. 2021. "Antimicrobial Activity of Pyrazinamide Coordination Frameworks Synthesized by Mechanochemistry" Molecules 26, no. 7: 1904. https://doi.org/10.3390/molecules26071904
APA StyleQuaresma, S., Alves, P. C., Rijo, P., Duarte, M. T., & André, V. (2021). Antimicrobial Activity of Pyrazinamide Coordination Frameworks Synthesized by Mechanochemistry. Molecules, 26(7), 1904. https://doi.org/10.3390/molecules26071904