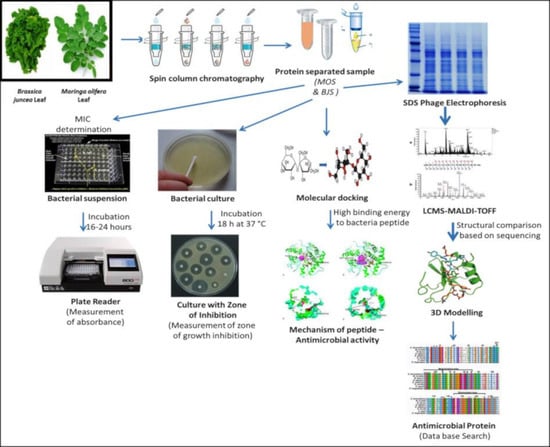
In Vitro and In Silico Screening and Characterization of Antimicrobial Napin Bioactive Protein in Brassica juncea and Moringa oleifera
Abstract
:1. Introduction
2. Results
2.1. Coagulation Activity and Protein Determination
2.2. Antibacterial Activity
2.2.1. Cell Aggregation
2.2.2. Determination of Minimum Inhibitory Concentration (MIC)
2.3. Identification of Bioactive Protein (Small Molecular Weight–Peptides) by Mass Spectrometry
2.4. Molecular Interaction of AM with Pathogens (Mode of Action)
SWISS-MODEL: Homology Modeling of Protein Structures
2.5. Antimicrobial Resistance
3. Discussion
4. Materials and Methods
4.1. Materials and Chemicals
4.2. Isolation of Pathogenic Microorganisms from Patient’s Sample
4.3. Microbial Strains Applied in the Study
4.4. Extraction of Crude Protein (Active Compounds) from Plant Leaf
4.5. Bioactive Protein—Functional Activity
4.5.1. Coagulation Activity
4.5.2. In Vitro Assay for Antimicrobial Activity
Disc Diffusion Kirby Bauer Method
Growth Inhibitory Activity—Determination of Minimum Inhibitory Activity (MIC) and Minimum Bactericidal Activity (MBC)
4.5.3. Electron Microscopy Analysis
4.6. Purification of Coagulant and Antimicrobial Protein
4.6.1. Purification of Protein Based on Column Chromatography
4.6.2. Tris-Tricine SDS-PAGE Electrophoresis and Gel-Elution
4.6.3. Quantification of Protein Concentration
4.6.4. Identification of Purified Peptides by Mass Spectrometry
4.7. SWISS-MODEL: Homology Modeling of Protein Structures
In Silico—Molecular Interaction Analysis and Docking for Antimicrobial Peptides in MOS
4.8. Antimicrobial Peptides (AMPs) with Limited Resistance
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Khurana, I.; Sen, R. WaterAid’s Vision Is of a World Where Everyone Has Access to Safe Water and Sanitation Drinking Water Quality in Rural India: Issues and Approaches. 2012. Available online: www.wateraid.org (accessed on 1 March 2021).
- Jepson, W.; Budds, J.; Eichelberger, L.; Harris, L.; Norman, E.; O’Reilly, K.; Stoler, J. Advancing human capabili-ties for water security: A relational approach. Water Secur. 2017, 1, 46–52. [Google Scholar] [CrossRef]
- Nduku, X. Effects of Dietary Inclusion of Moringa Oleifera Leaf Meal on Growth Performance, Physico-chemical Attrib-Utes, Oxidative Stability and Sensory Quality of Pork. Ph.D. Thesis, University of Fort Hare, Alice, Eastern cape, South Africa, April 2014. [Google Scholar]
- Bain, R.; Cronk, R.; Wright, J.; Yang, H.; Slaymaker, T.; Bartram, J. Fecal Contamination of Drinking-Water in Low- and Middle-Income Countries: A Systematic Review and Meta-Analysis. PLoS Med. 2014, 11, e1001644. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sunnen, G. A Primer of Water, Ozone and Health: The Challenge of Aquapollution; Ozonics International: New York, NY, USA, 2005. [Google Scholar]
- World Health Organization. Guidelines for Drinking-Water Quality. Available online: https://www.who.int/publications/i/item/9789241549950 (accessed on 1 March 2021).
- EPA, U. Primer for Municipal Wastewater Treatment Systems; EPA 832-R-04-001; EPA: Washington, DC, USA, 2004.
- Sato, T.; Qadir, M.; Yamamoto, S.; Endo, T.; Zahoor, A. Global, regional, and country level need for data on wastewater generation, treatment, and use. Agric. Water Manag. 2013, 130, 1–13. [Google Scholar] [CrossRef]
- Jed, W.; Fahey, S.D. Moringa oleifera: A Review of the Medical Evidence for Its Nutritional, Therapeutic, and Prophy-lactic Properties. Part 1 Trees Life J. 2005, 1, 5. [Google Scholar]
- Taiwo, A.S.; Adenike, K.; Aderonke, O. Efficacy of a natural coagulant protein from Moringa oleifera (Lam) seeds in treatment of Opa reservoir water, Ile-Ife, Nigeria. Heliyon 2020, 6, e03335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saleem, M.; Sami, A.J.; Bachmann, R.T. Characterisation and coagulant activity screening of fractionated wa-ter-soluble seed proteins from Moringa oleifera. Mater. Proc. 2020, 31, 207–210. [Google Scholar]
- Marobhe, N.J.; Dalhammar, G.; Gunaratna, K.R. Simple and rapid method for purification and characterization of ac-tive coagulants from the seeds of Vigna unguiculata and Parkinsonia aculeata. Environ. Technol. 2007, 28, 671–681. [Google Scholar] [CrossRef]
- Marobhe, N.J. Water Supply in Tanzania and Performance of Local Plant Materials in Purification of Turbid Water. TRI-TA-LWR. Ph.D. Thesis, KTH, School of Architecture and the Built Environment (ABE), Land and Water Resources Engineering. 1042, Stockholm, Sweden, June 2008. [Google Scholar]
- Futi, A.P.; Otieno, W.S.; Acholla, O.J.; Otieno, W.A.; Ochieng, O.S.; Mukisira, M.C. Moringa oleifera crude extracts and aluminum sulfate. J. Agri. Extn. Rural. Dev. 2011, 3, 102–112. [Google Scholar]
- Okunade, O.A.; Ghawi, S.K.; Methven, L.; Niranjan, K. Thermal and pressure stability of myrosinase enzymes from black mustard (Brassica nigra L. W.D.J. Koch. var. nigra), brown mustard (Brassica juncea L. Czern. var. juncea) and yellow mustard (Sinapsis alba L. subsp. maire) seeds. Food Chem. 2015, 187, 485–490. [Google Scholar] [CrossRef]
- Morra, M.J.; Borek, V. Glucosinolate preservation in stored Brassicaceae seed meals. J. Stored Prod. Res. 2010, 46, 98–102. [Google Scholar] [CrossRef]
- Gupta, S.; Jain, R.; Kachhwaha, S.; Kothari, S. Nutritional and medicinal applications of Moringa oleifera Lam.—Review of current status and future possibilities. J. Herb. Med. 2018, 11, 1–11. [Google Scholar] [CrossRef]
- Sousa, A.M.P.; Salles, H.O.; de Oliveira, H.D.; de Souza, B.B.P.; Filho, J.D.L.C.; Sifuentes, D.N.; Prates, M.V.; Junior, C.B.; Bemquerer, M.P.; Egito, A.S.D. Mo-HLPs: New flocculating agents identified from Moringa oleifera seeds belong to the hevein-like peptide family. J. Proteom. 2020, 217, 103692. [Google Scholar] [CrossRef] [PubMed]
- Lowry, O.H.; Rosebrough, N.J.; Farr, A.L.; Randall, R.J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar] [CrossRef]
- Schägger, H. Tricine–SDS-PAGE. Nat. Protoc. 2006, 1, 16–22. [Google Scholar] [CrossRef] [PubMed]
- Aluko, R.E.; McIntosh, T.; Katepa-Mupondwa, F. Comparative study of the polypeptide profiles and functional properties ofSinapis alba and Brassica juncea seed meals and protein concentrates. J. Sci. Food Agric. 2005, 85, 1931–1937. [Google Scholar] [CrossRef]
- Broin, M.; Santealla, C.; Cuine, S.; Kokou, K.; Peltier, G.; Joet, T. Flocculant activity of a recombinant protein from Moringa oleifera Lam. Seeds. Appl. Microbiol. Biotechnol. 2002, 60, 114–119. [Google Scholar]
- Kalibbala, H.M. Application of Indigenous Materials in Drinking Water Treatment. Ph.D. Thesis, KTH, School of Architecture and the Built Environment (ABE), Land and Water Resources Engineering, Stockholm, Sweden, 2007. [Google Scholar]
- Shetty, D.; Abrahante, J.E.; Chekabab, S.M.; Wu, X.; Korber, D.R.; Vidovic, S. Role of CpxR in Biofilm Development: Expression of Key Fimbrial, O-Antigen and Virulence Operons of Salmonella Enteritidis. Int. J. Mol. Sci. 2019, 20, 5146. [Google Scholar] [CrossRef] [Green Version]
- Lea, M. Bioremediation of Turbid Surface Water Using Seed Extract from Moringa oleifera Lam. (Drumstick) Tree. Curr. Protoc. Microbiol. 2010, 16, 1G.2.1–1G.2.14. [Google Scholar] [CrossRef]
- Wijetunge, D. Pathogenomics of neonatal meningitis causing Escherichia coli. Ph.D. Thesis, Pennsylvania State University, State College, PA, USA, March 2014. [Google Scholar]
- Olier, M. Escherichia coli: More Than A Pathogen? Probiot. Prebiot. Curr. Res. Futur. Trends 2015, 135, 135–152. [Google Scholar] [CrossRef] [Green Version]
- Silveira, F.M.R.; Baptista, A.T.A.; Dutra, T.V.; Filho, B.A.D.A.; Gomes, R.G.; Bergamasco, R. Application of Moringa oleifera Lam. fractionated proteins for inactivation of Escherichia coli from water. Water Sci. Technol. 2020, 81, 265–273. [Google Scholar] [CrossRef] [PubMed]
- Williams, F.E. The efficacy of Moringa oleifera as a practical application for sustainable water treatment. Ph.D. Thesis, The University of Adelaide, Adelaide, Australia, July 2019. [Google Scholar]
- D’Costa, V.M.; McGrann, K.M.; Hughes, D.W.; Wright, G.D. Sampling the Antibiotic Resistome. Science 2006, 311, 374–377. [Google Scholar] [CrossRef] [Green Version]
- Bacha, K.; Tariku, Y.; Gebreyesus, F.; Zerihun, S.; Mohammed, A.; Weiland-Bräuer, N.; Mulat, M. Antimicrobial and anti-Quorum Sensing activities of selected medicinal plants of Ethiopia: Implication for development of potent antimi-crobial agents. BMC Microbiol. 2016, 16, 139. [Google Scholar] [CrossRef] [Green Version]
- Martín-Rodríguez, A.J.; Quezada, H.; Becerril, G.; Maeda, V.P.; Wood, T.K.; García-Contreras, R. Recent advances in novel antibacterial development. Front Clin. Drug Res. Anti Infect. 2016, 2, 59. [Google Scholar]
- Cândido, E.D.S.; Pinto, M.F.S.; Pelegrini, P.B.; Lima, T.B.; Silva, O.N.; Pogue, R.; Grossi-De-Sá, M.F.; Franco, O.L. Plant storage proteins with antimicrobial activity: Novel insights into plant defense mechanisms. FASEB J. 2011, 25, 3290–3305. [Google Scholar] [CrossRef]
- Chandra, H.; Bishnoi, P.; Yadav, A.; Patni, B.; Mishra, A.P.; Nautiyal, A.R. Antimicrobial Resistance and the Al-ternative Resources with Special Emphasis on Plant-Based Antimicrobials—A Review. Plants 2017, 6, 16. [Google Scholar] [CrossRef] [PubMed]
- Nogueira, J.W.A.; Costa, R.A.; da Cunha, M.T.; Cavalcante, T.T.A. Antibiofilm activity of natural substances derived from plants. Afr. J. Microbiol. Res. 2017, 11, 1051–1060. [Google Scholar]
- Thevissen, K.; Kristensen, H.-H.; Thomma, B.P.; Cammue, B.P.; François, I.E. Therapeutic potential of antifungal plant and insect defensins. Drug Discov. Today 2007, 12, 966–971. [Google Scholar] [CrossRef] [PubMed]
- Geue, L.; Menge, C.; Eichhorn, I.; Semmler, T.; Wieler, L.H.; Pickard, D.; Berens, C.; Barth, S.A. Evidence for Contemporary Switching of the O-Antigen Gene Cluster between Shiga Toxin-Producing Escherichia coli Strains Colonizing Cattle. Front. Microbiol. 2017, 8, 424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gast, R.K.; Porter, R.E., Jr. Salmonella infections. In Diseases of Poultry Chapter 16; Swayne, D.E., Boulianne, M., Logue, C.M., McDougald, L.R., Nair, V., Suarez, D.L., Wit, S., Grimes, T., Johnson, D., Kromm, M., Eds.; John Willey & Sons Inc.: Hoboken, NJ, USA, 2020; pp. 717–753. [Google Scholar]
- McClelland, M.; Sanderson, K.E.; Spieth, J.; Clifton, S.W.; Latreille, P.; Courtney, L.; Porwollik, S.; Ali, J.; Dante, M.; Du, F.; et al. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nat. Cell Biol. 2001, 413, 852–856. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McClelland, M.; Sanderson, K.E.; Clifton, S.W.; Latreille, P.; Porwollik, S.; Sabo, A.; Meyer, R.; Bieri, T.; Ozersky, P.; Harkins, C.R.; et al. Comparison of genome degradation in Paratyphi A and Typhi, human-restricted serovars of Salmonella enterica that cause typhoid. Nat. Genet. 2004, 36, 1268–1274. [Google Scholar] [CrossRef]
- Keusch, G.T.; Grady, G.F.; Mata, L.J.; McIver, J. The pathogenesis of Shigella Diarrhea: I. Enterotoxin production by Shigella dysenteriae 1. J. Clin. Investig. 1972, 51, 1212–1218. [Google Scholar] [CrossRef] [PubMed]
- Adekoya, I.; Obadina, A.; Olorunfemi, M.; Akande, O.; Landschoot, S.; De Saeger, S.; Njobeh, P. Occurrence of bacteria and endotoxins in fermented foods and beverages from Nigeria and South Africa. Int. J. Food Microbiol. 2019, 305, 108251. [Google Scholar] [CrossRef] [PubMed]
- Harrison, L.M.; Gaines, D.W.; Babu, U.S.; Balan, K.V.; Reimschuessel, R.; Do, A.B.; Williams, K.M. Di-et-induced obesity precipitates kidney dysfunction and alters inflammatory mediators in mice treated with Shiga toxin 2. Microbial. Pathog. 2018, 123, 250–258. [Google Scholar] [CrossRef]
- Tsirigotaki, A.; De Geyter, J.; Šoštaric, N.; Economou, A.; Karamanou, A.T.J.D.G.N.; Šoštarić, A.E.S. Protein export through the bacterial Sec pathway. Nat. Rev. Genet. 2017, 15, 21–36. [Google Scholar] [CrossRef]
- Nelson, M.; Grier, M.; Barbaro, S.; Ismail, M. Polyfunctional Antibiotics Affecting Bacterial Membrane Dynamics. Anti-Infect. Agents Med. Chem. 2009, 8, 3–16. [Google Scholar] [CrossRef]
- Panteleev, P.V.; Myshkin, M.Y.; Shenkarev, Z.O.; Ovchinnikova, T.V. Dimerization of the antimicrobial peptide arenicin plays a key role in the cytotoxicity but not in the antibacterial activity. Biochem. Biophys. Res. Commun. 2017, 482, 1320–1326. [Google Scholar] [CrossRef] [PubMed]
- Dahot, M. Antibacterial and antifungal activity of small protein of Indigofera oblongifolia leaves. J. Ethnopharmacol. 1999, 64, 277–282. [Google Scholar] [CrossRef]
- Shenkarev, Z.O.; Balandin, S.V.; Trunov, K.I.; Paramonov, A.S.; Sukhanov, S.V.; Barsukov, L.I.; Ovchinnikova, T.V. Molecular mechanism of action of β-hairpin antimicrobial peptide arenicin: Oligomeric structure in dodecylphospho-choline micelles and pore formation in planar lipid bilayers. Biochemistry 2011, 50, 6255–6265. [Google Scholar] [CrossRef]
- Hald, C.; Dawid, C.; Tressel, R.; Hofmann, T. Kaempferol 3-O-(2‴-O-Sinapoyl-β-sophoroside) Causes the Undesired Bitter Taste of Canola/Rapeseed Protein Isolates. J. Agric. Food Chem. 2018, 67, 372–378. [Google Scholar] [CrossRef]
- Pinto, C.E.M.; Farias, D.F.; Carvalho, A.F.U.; Oliveira, J.T.; Pereira, M.L.; Grangeiro, T.B.; Freire, J.E.; Viana, D.A.; Vasconcelos, I.M. Food safety assessment of an antifungal protein from Moringa oleifera seeds in an agricultural biotechnology perspective. Food Chem. Toxicol. 2015, 83, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Chelliah, R.; Saravanakumar, K.; Daliri, E.B.-M.; Kim, J.-H.; Lee, J.-K.; Jo, H.-Y.; Madar, I.H.; Kim, S.-H.; Ramakrishnan, S.R.; Rubab, M.; et al. An effective datasets describing antimicrobial peptide produced from Pediococcus acidilactici-purification and mode of action determined by molecular docking. Data Brief 2020, 31, 105745. [Google Scholar] [CrossRef] [PubMed]
- Kozakov, D.; Hall, D.R.; Xia, B.; Porter, K.A.; Padhorny, D.; Yueh, C.; Beglov, D.; Vajda, S. The ClusPro web server for protein-protein docking. Nat. Protoc. 2017, 12, 255–278. [Google Scholar] [CrossRef]
- Tsodikov, O.V.; Record, M.T.; Sergeev, Y.V. Novel computer program for fast exact calculation of accessible and molecular surface areas and average surface curvature. J. Comput. Chem. 2002, 23, 600–609. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, R.A.; Swindells, M.B. Ligplot+: Multiple Ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 2011, 51, 2778–2786. [Google Scholar] [CrossRef] [PubMed]
- Okada, K. Is Omega Squared Less Biased? A Comparison of Three Major Effect Size Indices in One-Way Anova. Behaviour 2013, 40, 129–147. [Google Scholar] [CrossRef]





| SNo | List of Microorganism | Minimum Inhibitory Concentration (MIC) | |||
|---|---|---|---|---|---|
| Moringaolifera Leaf | Brassica juncea Leaf | ||||
| Crude Protein (MOS) (mg/mL) | Coagulant Protein (MCP) (mg/mL) | Crude Protein (BJS) (mg/mL) | Coagulant Protein (BJP) (mg/mL) | ||
| 1 | E. coli | 0.01 | 0.02 | 0.05 | 0.09 |
| 2 | E. faecalis | 0.01 | 0.02 | 0.02 | 0.04 |
| 3 | S. paratyphi B | 0.04 | 0.06 | 0.05 | 0.09 |
| 4 | E. species | 0.01 | 0.02 | 0.05 | 0.08 |
| 5 | S. flexneri | 0.02 | 0.04 | 0.04 | 0.07 |
| 6 | P. mirabilis | 0.008 | 0.01 | 0.05 | 0.07 |
| 7 | S. aureus | 0.01 | 0.02 | 0.05 | 0.09 |
| 8 | S. typhimurium | 0.04 | 0.06 | 0.03 | 0.06 |
| 9 | S. typhi | 0.02 | 0.04 | 0.05 | 0.08 |
| 10 | S. marcescens | 0.01 | 0.02 | 0.05 | 0.08 |
| 11 | K. pneumoniae | 0.01 | 0.02 | 0.05 | 0.09 |
| 12 | S. paratyphi A | 0.02 | 0.04 | 0.02 | 0.04 |
| 13 | S. dysenteriae | 0.01 | 0.02 | 0.05 | 0.08 |
| Type Of Interactions | Napin | 3u1y | Total | NPN-3U1Y | Total-MSA | Weighted Score | Representative |
| Napin_3u1y | 2768.19 | 25424.35 | 28192.54 | 25088.38 | 3104.16 | −786 | Lowest Energy |
| Napin | 4plb | Total | NPN-4PLB | Total-MSA | Weighted Score | Representative | |
| Napin_4plb | 2768.19 | 54046.17 | 56814.36 | 53812.11 | 3002.25 | −912.9 | Lowest Energy |
| Type of Interactions | Hydrogen | Hydrophobic | ||
|---|---|---|---|---|
| Chain ID | Chain A (Napin) | Chain B (3U1Y) | Chain A (Napin) | Chain B (3U1Y) |
| 3U1Y | GLU 12, GLU 8 GLN 24, TRP 21 SER 29, ALA 27 PRO 30, ARG 23 GLN 19, HIS 14 GLN 25, GLN 10 | ARG 465, LYS 441, ASP 458, THR 360, GLN 502, LYS 371, TYR 498, ASP 495, ARG 494, GLU 518, HIS 563, LYS 537, ASN 519 | EU 15, GLN 6 CYS 18, PHE 9 ILE 22, LEU 26 GLY 28 | ASP 460,PRO 462, LYS 560, PHE 459, PHE 4922, GLY 491, MET 361, LEU 499, LEU 504 |
| Chain ID | Chain A (Napin) | Chain B (4PLB) | Chain A (Napin) | Chain B (4PLB) |
| 4PLB | PHE 31, PRO 30, GLN 1, GLN 6, GLN 25, GLN 20, CYS 18, LYS 4, GLU 8, CYS 5 | ALA 640, LYS 607, ALA 534, ASN 1010, GLU 1011, SER 1021, ASP 1024, ARG 517, THR 544, THR 594, GLU 599, ARG 601, LYS 502 | GLU 12, ILE 22, PHE 9, LEU 26, GLN 24, TRP 21, PRO 2 | GLY 1341, VAL 1031, MET 1027, ARG 1012, TYR 639, HIS 600, GLN 505, GLU 1020, LEU 603, ALA 602, ALA 509, PRO 542, ILE 539, GLN 541, HIS 501, PRO 598 |
| SNo | List of Pathogens | Nucleic Acid Targeting Antibiotic (A) | Cell Wall Targeting Antibiotic (B) | Protein Synthesis Inhibiting Targeting Antibiotic (C) | ||
|---|---|---|---|---|---|---|
| Novobiocin (mm) | Clindamycin (mm) | Gentamicin (mm) | Ampicillin (mm) | Vancomycin (mm) | ||
| 1 | E. coli | 12.00 ± 0.05 c | 14.00 ± 0.10 b | 16.00 ± 0.05 b | 12.00 ± 0.07 c | 13.00 ± 0.05 c |
| 2 | E. faecalis | 14.00 ± 0.03 a | - | 17.00 ± 0.02 b | 12.00 ± 0.01 c | 12.00 ± 0.03 c |
| 3 | S. paratyphi B | - | - | - | 16.00 ± 0.01 b | 10.00 ± 0.07 c |
| 4 | E. species | - | - | 13.00 ± 0.05 c | 14.00 ± 0.03 b | 15.00 ± 0.03 b |
| 5 | S. flexneri | - | - | 15.00 ± 0.03 b | 15.00 ± 0.04 a | 17.00 ± 0.03 b |
| 6 | P. mirabilis | - | 17.00 ± 0.03 b | 16.00 ± 0.03 b | 14.00 ± 0.11 b | |
| 7 | S. aureus | - | - | 14.00 ± 0.08 b | 15.00 ± 0.02 b | |
| 8 | S. typhimurium | - | - | 15.00 ± 0.03 b | - | |
| 9 | S. typhi | 16.00 ± 0.05 a | 17.00 ± 0.02 a | 12.00 ± 0.03 c | 14.00 ± 0.09 b | 12.00 ± 0.03 c |
| 10 | S. marcescens | 12.00 ± 0.12 c | 15.00 ± 0.06 a | 15.00 ± 0.03 b | 12.00 ± 0.04 c | 10.00 ± 0.02 c |
| 11 | K. pneumoniae | - | - | - | 12.00 ± 0.03 c | 13.00 ± 0.05 c |
| 12 | S. paratyphi A | - | - | - | 14.00 ± 0.05 b | 12.00 ± 0.01 c |
| 13 | S. dysenteriae | - | 14.00 ± 0.04 b | 14.00 ± 0.03 c | 10.00 ± 0.02 c | 10.00 ± 0.06 c |
| Sample SNo: | List of Pathogens | Moringa olifera (MOS) | Brassica juncea (BJS) | ||
|---|---|---|---|---|---|
| Aggregation | Growth Inhibition | Aggregation | Growth Inhibition | ||
| 1 | E. coli | + | + | + | + |
| 2 | E. faecalis | + | + | + | + |
| 3 | S. paratyphi B | + | - | + | - |
| 4 | E. species | + | + | + | - |
| 5 | S. flexneri | + | + | + | + |
| 6 | P. mirabilis | + | + | + | - |
| 7 | S. aureus | + | + | + | - |
| 8 | S. typhimurium | + | - | - | - |
| 9 | S. typhi | + | + | - | - |
| 10 | S. marcescens | + | - | - | - |
| 11 | K. pneumoniae | - | + | + | - |
| 12 | S. paratyphi A | - | + | - | + |
| 13 | S. dysenteriae | - | + | - | + |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chandrashekar, S.; Vijayakumar, R.; Chelliah, R.; Daliri, E.B.-M.; Madar, I.H.; Sultan, G.; Rubab, M.; Elahi, F.; Yeon, S.-J.; Oh, D.-H. In Vitro and In Silico Screening and Characterization of Antimicrobial Napin Bioactive Protein in Brassica juncea and Moringa oleifera. Molecules 2021, 26, 2080. https://doi.org/10.3390/molecules26072080
Chandrashekar S, Vijayakumar R, Chelliah R, Daliri EB-M, Madar IH, Sultan G, Rubab M, Elahi F, Yeon S-J, Oh D-H. In Vitro and In Silico Screening and Characterization of Antimicrobial Napin Bioactive Protein in Brassica juncea and Moringa oleifera. Molecules. 2021; 26(7):2080. https://doi.org/10.3390/molecules26072080
Chicago/Turabian StyleChandrashekar, Sangeeta, Raman Vijayakumar, Ramachandran Chelliah, Eric Banan-Mwine Daliri, Inamul Hasan Madar, Ghazala Sultan, Momna Rubab, Fazle Elahi, Su-Jung Yeon, and Deog-Hwan Oh. 2021. "In Vitro and In Silico Screening and Characterization of Antimicrobial Napin Bioactive Protein in Brassica juncea and Moringa oleifera" Molecules 26, no. 7: 2080. https://doi.org/10.3390/molecules26072080
APA StyleChandrashekar, S., Vijayakumar, R., Chelliah, R., Daliri, E. B.-M., Madar, I. H., Sultan, G., Rubab, M., Elahi, F., Yeon, S.-J., & Oh, D.-H. (2021). In Vitro and In Silico Screening and Characterization of Antimicrobial Napin Bioactive Protein in Brassica juncea and Moringa oleifera. Molecules, 26(7), 2080. https://doi.org/10.3390/molecules26072080