Synthesis, Biomacromolecular Interactions, Photodynamic NO Releasing and Cellular Imaging of Two [RuCl(qn)(Lbpy)(NO)]X Complexes
Abstract
:1. Introduction
2. Results and Discussion
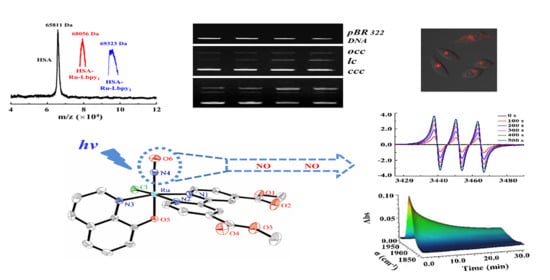
2.1. Synthesis and Characterization
2.2. Photo-Induced NO Release
2.3. Protein Binding Properties
2.4. DNA Binding Properties
2.5. Cytotoxicity and Imaging of NO Releasing in Living Cell
3. Materials and Methods
3.1. Materials
3.2. Measurements
3.3. Synthesis
3.3.1. Synthesis of [RuCl(qn)(Lbpy1)(NO)]Cl (Ru-Lbpy1)
3.3.2. Synthesis of [RuCl(qn)(Lbpy2)(NO)]NO3 (Ru-Lbpy2)
3.4. Photo-Induced NO Release
3.5. Binding of the Complexes with Protein
3.6. Binding of the Complexes with DNA and Photo-Induced DNA Damage
3.7. Cellular Culture
3.8. Cellular Imaging of NO
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Ignarro, L.J.; Freeman, B.A. Nitric oxide: Biology and Pathobiology, 3rd ed.; Academic Press: Cambridge, MA, USA, 2017. [Google Scholar]
- Ataabadi, E.A.; Golshiri, K.; Jüttner, A.; Krenning, G.; Danser, A.H.J.; Roks, A.J.M. Nitric oxide-cGMP signaling in hypertension: Current and future options for pharmacotherapy. Hypertension 2020, 76, 1055–1068. [Google Scholar] [CrossRef]
- Tewari, D.; Sah, A.N.; Bawari, S.; Nabavi, S.F.; Dehpour, A.R.; Shirooie, S.; Braidy, N.; Fiebich, B.L.; Vacca, R.A.; Nabavi, S.M. Role of nitric oxide in neurodegeneration: Function, regulation and inhibition. Curr. Neuropharmacol. 2021, 19, 114–126. [Google Scholar] [CrossRef]
- Gantner, B.N.; LaFond, K.M.; Bonini, M.G. Nitric oxide in cellular adaptation and disease. Redox. Biol. 2020, 34. [Google Scholar] [CrossRef]
- Lisi, F.; Zelikin, A.N.; Chandrawati, R. Nitric oxide to fight viral infections. Adv. Sci. 2021, 8, 2003895. [Google Scholar] [CrossRef] [PubMed]
- Dimmeler, S.; Zeihar, A.M. Nitric oxide and apoptosis: Another paradigm for the double-edged role of nitric oxide. Nitric Oxide 1997, 1, 275–281. [Google Scholar] [CrossRef] [PubMed]
- Mocellin, S.; Bronte, V.; Nitti, D. Nitric oxide, a double edged sword in cancer biology: Searching for therapeutic opportunities. Med. Res. Rev. 2007, 27, 317–352. [Google Scholar] [CrossRef] [PubMed]
- Boyd, C.S.; Cadenas, E. Nitric oxide and cell signaling pathways in mitochondrial-dependent apoptosis. Biol. Chem. 2002, 383, 411–423. [Google Scholar] [CrossRef] [PubMed]
- Brown, G.C. Regulation of mitochondrial respiration by nitric oxide inhibition of cytochrome c oxidase. Biochim. Biophys. Acta 2001, 1504, 46–57. [Google Scholar] [CrossRef] [Green Version]
- Payne, C.M.; Waltmire, C.N.; Crowley, C.; Crowley-Weber, C.L.; Dvorakova, K.; Bernstein, H.; Bernstein, C.; Holubec, H.; Garewal, H. Caspase-6 mediated cleavage of guanylate cyclase alpha 1 during deoxycholate-induced apoptosis protective role of the nitric oxide signaling module. Cell. Biol. Toxicol. 2003, 19, 373–392. [Google Scholar] [CrossRef]
- Chung, P.; Cook, T.; Liu, K.; Vodovotz, Y.; Zamora, R.; Finkelstein, S.; Billiar, T.; Blumberg, D. Overexpression of the human inducible nitric oxide synthase gene enhances radiation-induced apoptosis in colorectal cancer cells via a caspase-dependent mechanism. Nitric Oxide 2003, 8, 119–126. [Google Scholar] [CrossRef]
- Bradley, S.A.; Steinert, J.R. Nitric oxide-mediated posttranslational modifications: Impacts at the synapse. Oxi. Med. Cell. Longev. 2016, 2016, 5681036. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dyer, R.R.; Ford, K.I.; Robinson, R.A.S. The roles of S-nitrosylation and S-glutathionylation in Alzheimer’s disease. Meth. Enzymol. 2019, 626, 499–538. [Google Scholar]
- Rose, M.J.; Mascharak, P.K. Photoactive ruthenium nitrosyls: Effects of light and potential application as NO donors. Coord. Chem. Rev. 2008, 252, 2093–2114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rose, M.J.; Fry, N.L.; Marlow, R.; Hinck, L.; Mascharak, P.K. Sensitization of ruthenium nitrosyls to visible light via direct coordination of the dye resorufin: Trackable NO donors for light-triggered NO delivery to cellular targets. J. Am. Chem. Soc. 2008, 130, 8834–8846. [Google Scholar] [CrossRef] [PubMed]
- Patra, A.K.; Mascharak, P.K. A ruthenium nitrosyl that rapidly delivers NO to proteins in aqueous solution upon short exposure to UV light. Inorg. Chem. 2003, 42, 7363–7365. [Google Scholar] [CrossRef]
- Awasabisah, D.; Xu, N.; Gautam, K.P.S.; Powell, D.R.; Shaw, M.J.; Richter-Addo, G.B. Preparation, characterization, electrochemistry, and infrared spectroelectrochemistry of ruthenium nitrosyl porphyrins containing η1-O-bonded axial carboxylates. Eur. J. Inorg. Chem. 2016, 2016, 509–518. [Google Scholar] [CrossRef] [Green Version]
- Song, L.N.; Xie, L.L.; Xu, L.Q.; Jing, Q.; Liu, C.Y.; Xi, X.L.; Wang, W.M.; Zhao, Y.; Zhao, X.A.; Wang, H.F. Syntheses, spectra, photoinduced nitric oxide release and interactions with biomacromolecules of three nitrosylruthenium complexes. Polyhedron 2020, 185, 114596. [Google Scholar] [CrossRef]
- Hirano, T.; Oi, T.; Nagao, H.; Morokuma, K. Regulation of geometry around the ruthenium center of bis(2-pyridinecarboxylato) complexes by the nitrosyl moiety: Syntheses, structures, and theoretical studies. Inorg. Chem. 2003, 42, 6575–6583. [Google Scholar] [CrossRef]
- Gomes, A.J.; Espreafico, E.M.; Tfouni, E. Trans-[Ru(NO)Cl(cyclam)](PF6)2 and [Ru(NO)(Hedta)] incorporated in PLGA nanoparticles for the delivery of nitric oxide to B16-F10 cells: Cytotoxicitity and phototoxicity. Mol. Pharmaceut. 2013, 10, 3544–3554. [Google Scholar] [CrossRef]
- Prakash, R.; Czaja, A.U.; Heinemann, F.W.; Sellmann, D. Visible light induced reversible extrusion of nitric oxide from a ruthenium(II) nitrosyl complex: A facile delivery of nitric oxide. J. Am. Chem. Soc. 2005, 127, 13758–13759. [Google Scholar] [CrossRef]
- Shoair, A.F.; El-Bindary, A.A. Synthesis, spectral and catalytic dehydrogenation studies of ruthenium complexes containing NO bidentate ligands. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2014, 131, 490–496. [Google Scholar] [CrossRef] [PubMed]
- Cherdtrakulkiat, R.; Boonpangrak, S.; Sinthupoom, N.; Prachayasittikul, S.; Ruchirawat, S.; Prachayasittikul, V. Derivatives (halogen, nitro and amino) of 8-hydroxyquinoline with highly potent antimicrobial and antioxidant activities. Biochem. Biophys. Rep. 2016, 6, 135–141. [Google Scholar] [CrossRef] [Green Version]
- Oliveri, V.; Vecchio, G. 8-Hydroxyquinolines in medicinal chemistry: A structural perspective. Eur. J. Med. Chem. 2016, 120, 252–274. [Google Scholar] [CrossRef]
- Heinemann, F.; Karges, J.; Gasser, G. Critical overview of the use of Ru(II) polypyridyl complexes as photosensitizers in one-photon and two-photon photodynamic therapy. Acc. Chem. Res. 2017, 50, 2727–2736. [Google Scholar] [CrossRef] [PubMed]
- Thota, S.; Rodrigues, D.A.; Crans, D.C.; Barreiro, E.J. Ru(II) Compounds: Next-Generation Anticancer Metallotherapeutics? J. Med. Chem. 2018, 61, 5805–5821. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhu, K.X.; Cao, L.; Xie, Z.F.; Gu, M.; Lü, W.; Li, J.Y.; Nan, F.J. Berberine derivatives with a long alkyl chain branched by hydroxyl group and methoxycarbonyl group at 9-position show improved anti-proliferation activity and membrane permeability in A549 cells. Acta. Pharmacol. Sin. 2020, 41, 813–824. [Google Scholar] [CrossRef]
- Mori, R.; Kato, A.; Komenoi, K.; Kurasaki, H.; Iijima, T.; Kawagoshi, M.; Kiran, Y.B.; Takeda, S.; Sakai, N.; Konakahara, T. Synthesis and in vitro antitumor activity of novel 2-alkyl-5-methoxycarbonyl-11-methyl-6H-pyrido[4,3-b]carbazol-2-ium and 2-alkylellipticin-2-ium chloride derivatives. Eur. J. Med. Chem. 2014, 82, 16–35. [Google Scholar] [CrossRef]
- Gu, Y.; Lee, H.M.; Napolitano, N.; Clemens, M.; Zhang, Y.Z.; Sorsa, T.; Zhang, Y.; Johnson, F.; Golub, L.M. 4-methoxycarbonyl curcumin: A unique inhibitor of both inflammatory mediators and periodontal inflammation. Mediat. Inflamm. 2013, 2013, 329740. [Google Scholar] [CrossRef]
- Merkle, A.C.; McQuarters, A.B.; Lehnert, N. Synthesis, spectroscopic analysis and photolabilization of water-soluble ruthenium(III)-nitrosyl complexes. Dalton. Trans. 2012, 41, 8047–8059. [Google Scholar] [CrossRef] [PubMed]
- Roose, M.; Tassé, M.; Lacroix, P.G.; Malfant, I. Nitric oxide (NO) photo-release in a series of ruthenium-nitrosyl complexes: New experimental insights in the search for a comprehensive mechanism. New. J. Chem. 2019, 43, 755–767. [Google Scholar] [CrossRef]
- Labra-Vázquez, P.; Bocé, M.; Tassé, M.; Mallet-Ladeira, S.; Lacroix, P.G.; Farfán, N.; Malfant, I. Chemical and photochemical behavior of ruthenium nitrosyl complexes with terpyridine ligands in aqueous media. Dalton. Trans. 2020, 49, 138–3154. [Google Scholar] [CrossRef]
- Napoli, C.; Paolisso, G.; Casamassimi, A.; Al-Omran, M.; Barbieri, M.; Sommese, L.; Infante, T.; Ignarro, L.J. Effects of nitric oxide on cell proliferation: Novel insights. J. Am. Coll. Cardiol. 2013, 62, 89–95. [Google Scholar] [CrossRef]
- Burke, A.J.; Sullivan, F.J.; Giles, F.J.; Glynn, S.A. The yin and yang of nitric oxide in cancer progression. Carcinogenesis 2013, 34, 503–512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vanin, A.F.; Poltorakov, A.P.; Mikoyan, V.D.; Kubrina, L.N.; van Faassen, E. Why iron-dithiocarbamates ensure detection of nitric oxide in cells and tissues. Nitric Oxide 2006, 15, 295–311. [Google Scholar]
- Patra, A.K.; Rose, M.J.; Murphy, K.A.; Olmstead, M.M.; Mascharak, P.K. Photolabile ruthenium nitrosyls with planar dicarboxamide tetradentate N4 Ligands: Effects of in-plane and axial ligand strength on NO release. Inorg. Chem. 2004, 43, 4487–4495. [Google Scholar] [CrossRef]
- Nataliia, M.; Pascal, G.L.; Valerii, B.; Marine, T.; Carine, D.; Martial, B.P.; Isabelle, M. Multistep photochemical reactions of polypyridine-based ruthenium nitrosyl complexes in dimethylsulfoxide. Molecules 2020, 25, 2205. [Google Scholar]
- Pou, S.; Tsai, P.; Porasuphatana, S.; Halpern, H.J.; Chandramouli, G.V.R.; Barth, E.D.; Rosen, G.M. Spin trapping of nitric oxide by ferro-chelates: Kinetic and in vivo pharmacokinetic studies. Biochim. Biophys. Acta 1999, 1427, 216–226. [Google Scholar] [CrossRef]
- Weaver, J.; Porasuphatana, S.; Budzichowski, T.; Rosen, G.M. Spin trapping nitric oxide from neuronal nitric oxide synthase: A look at several iron- dithiocarbamate complexes. Free Radical Res. 2005, 39, 1027–1033. [Google Scholar] [CrossRef]
- Dömötör, O.; Rathgeb, A.; Kuhn, P.S.; Popović-Bijelić, A.; Bačić, G.; Enyed, E.A.; Arion, V.B. Investigation of the binding of cis/trans-[MCl4(1H-indazole)(NO)]- (M = Ru, Os) complexes to human serum albumin. J. Inorg. Biochem. 2016, 159, 37–44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Curry, S.; Mandelkow, H.; Brick, P.; Franks, N. Crystal structure of human serum albumin complexed with fatty acid reveals an asymmetric distribution of binding sites. Nat. Struct. Mol. Biol. 1998, 5, 827–835. [Google Scholar] [CrossRef]
- Ito, S.; Senoo, A.; Nagatoishi, S.; Yamamoto, M.; Tsumoto, K.; Wakui, N. Structural basis for the binding mechanism of human serum albumin complexed with cyclic peptide dalbavancin. J. Med. Chem. 2020, 63, 14045–14053. [Google Scholar] [CrossRef]
- Gill, M.R.; Thomas, J.A. Ruthenium(II) polypyridyl complexes and DNA-from structural probes to cellular imaging and therapeutics. Chem. Soc. Rev. 2012, 41, 3179–3192. [Google Scholar] [CrossRef]
- Liu, Y.; Li, Z.; Liang, Z.; Yao, J.; Huang, H. Cytotoxicity, apoptosis, cellular uptake, cell cycle arrest, photocleavage, and antioxidant activity of 1,10-phenanthroline ruthenium(II) complexes. DNA Cell Bio. 2011, 30, 839–848. [Google Scholar] [CrossRef]
- Teixeira, T.M.; Arraes, I.G.; Abreu, D.C.; Oliveira, K.M.; Correa, R.S.; Batista, A.A.; Braunbeck, T.; Lacerda, E.D.P.S. Ruthenium complexes show promise when submitted to toxicological safety tests using alternative methodologies. Eur. J. Med. Chem. 2021, 216, 113262. [Google Scholar] [CrossRef]
- Xiong, K.; Qian, C.; Yuan, Y.; Wei, L.; Liao, X.; He, L.; Rees, T.W.; Chen, Y.; Wan, J.; Ji, L.; et al. Necroptosis induced by ruthenium(II) complexes as dual catalytic inhibitors of topoisomerase I/II. Angew. Chem. Int. Edit. 2020, 59, 16631–16637. [Google Scholar] [CrossRef] [PubMed]
- Maji, M.; Acharya, S.; Maji, S.; Purkait, K.; Gupta, A.; Mukherjee, A. Differences in stability, cytotoxicity, and mechanism of action of Ru(II) and Pt(II) complexes of a bidentate N,O donor ligand. Inorg. Chem. 2020, 59, 10262–10274. [Google Scholar] [CrossRef] [PubMed]
- Hager, L.A.; Mokesch, S.; Kieler, C.; Castro, A.D.S.; Baier, D.; Roller, A.; Kandioller, W.; Keppler, B.K.; Berger, W.; Salassa, L.; et al. Ruthenium-arene complexes bearing naphthyl-substituted 1,3-dioxoindan-2-carboxamides ligands for G-quadruplex DNA recognition. Dalton. Trans. 2019, 48, 12040–12049. [Google Scholar] [CrossRef] [Green Version]
- Savic, M.; Arsenijevic, A.; Milovanovic, J.; Stojanovic, B.; Stankovic, V.; Simovic, R.A.; Lazic, D.; Arsenijevic, N.; Milovanovic, M. Antitumor activity of ruthenium(II) terpyridine complexes towards colon cancer cells in vitro and in vivo. Molecules 2020, 25, 4699. [Google Scholar] [CrossRef]
- Renfrew, A.K.; Karges, J.; Scopelliti, R.; Bobbink, F.D.; Nowak-Sliwinska, P.; Gasser, G.; Dyson, P.J. Towards light-activated ruthenium-arene (RAPTA-type) prodrug candidates. Chembiochem 2019, 20, 2876–2882. [Google Scholar] [CrossRef]
- Notaro, A.; Frei, A.; Rubbiani, R.; Jakubaszek, M.; Basu, U.; Koch, S.; Mari, C.; Dotou, M.; Blacque, O.; Gouyon, J.; et al. Ruthenium(II) complex containing a redox-active semiquinonate ligand as a potential chemotherapeutic agent: From synthesis to in vivo studies. J. Med. Chem. 2020, 63, 5568–5584. [Google Scholar] [CrossRef]
- Liu, J.G.; Lai, H.Q.; Xiong, Z.S.; Chen, B.L.; Chen, T.F. Functionalization and cancer-targeting design of ruthenium complexes for precise cancer therapy. Chem. Commun. 2019, 55, 9904–9914. [Google Scholar] [CrossRef] [PubMed]
- Pettinari, R.; Marchetti, F.; Tombesi, A.; Duan, F.H.; Zhou, L.M.; Messori, L.; Giacomelli, C.; Marchetti, L.; Trincavelli, M.L.; Marzo, T.; et al. Ruthenium(II) 1,4,7-trithiacyclononane complexes of curcumin and bisdemethoxycurcumin: Synthesis, characterization, and biological activity. J. Inorg. Biochem. 2021, 218, 111387. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.F.; Hagihara, T.; Ikezawa, H.; Tomizawa, H.; Miki, E. Electronic effects of the substituent group in 8-quinolinolato ligand on geometrical isomerism for nitrosylruthenium(II) complexes. Inorg. Chim. Acta 2000, 299, 80–90. [Google Scholar] [CrossRef]
- Liu, K.Y.; Ko, C.Y.; Ho, K.C.; Lin, K.F. Synthesis and characterization of cross-linkable ruthenium dye with ion coordinating property for dye-sensitized solar cells. Polymer 2011, 52, 3318–3324. [Google Scholar] [CrossRef]
- Wang, J.R.; Yang, F.; Zhao, Y.; Yu, P.Y.; Qiao, X.Y.; Wang, J.P.; Wang, H.F. Photoisomerization and structural dynamics of two nitrosylruthenium complexes: A joint study by NMR and nonlinear IR spectroscopies. Phys. Chem. Chem. Phys. 2014, 16, 24045–24054. [Google Scholar] [CrossRef]










| Chemical formula | C25H24ClN5O10Ru | α (°) | 105.601(2) |
| Formula weight | 691.01 | β (°) | 101.476(3) |
| Temperature (K) | 296 | γ (°) | 107.325(2) |
| Color | brown | Volume (Å3) | 1390.9(2) |
| Size (mm) | 0.35 × 0.35 × 0.33 | Z | 2 |
| Crystal system | Triclinic | Calculated density Dx g/cm−3 | 1.650 |
| Space group a (Å) b (Å) c (Å) | P-1 10.5852(9) 11.4031(10) 13.1528(12) | R [F2 > 2σ(F2)] wR2 (F2) GOOF CCDC number | 0.0663 0.1663 1.023 2054814 |
| Ru1—Cl1 | 2.373(2) | N1—Ru1—N2 | 79.3(2) |
| Ru1—N1 | 2.068(5) | N1—Ru1—N3 | 166.6(2) |
| Ru1—N2 | 2.073(6) | N1—Ru1—N4 | 95.9(3) |
| Ru1—N3 | 2.070(5) | N1—Ru1—O5 | 85.9(2) |
| Ru1—N4 | 1.752(6) | N2—Ru1—N3 | 95.6 (2) |
| Ru1—O5 | 1.980(4) | N2—Ru1—N4 | 92.4(3) |
| N4—O6 | 1.101(9) | N2—Ru1—O5 | 84.8(2) |
| Cl1—Ru1—N1 | 96.0(2) | N3—Ru1—N4 | 96.7(3) |
| Cl1—Ru1—N2 | 172.0(2) | N3—Ru1—O5 | 81.3(2) |
| Cl1—Ru1—N3 | 87.6(2) | N4—Ru1—O5 | 176.4(2) |
| Cl1—Ru1—N4 | 94.4(2) | Ru1—N4—O6 | 177.6(6) |
| Cl1—Ru1—O5 | 88.5(1) |
| HSA | DNA | |||
|---|---|---|---|---|
| Complex | Kb/(L·mol−1) | n | Kb/(L·mol−1) | n |
| Ru-bpy1 | 4.83 ± 0.09 × 106 | 1.46 | 2.14 ± 0.08 × 104 | 1.20 |
| Ru-bpy2 | 7.41 ± 0.08 × 104 | 1.09 | 5.52 ± 0.16 × 103 | 1.14 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, L.; Bai, H.; Liu, C.; Gong, W.; Wang, A.; Wang, L.; Zhao, Y.; Zhao, X.; Wang, H. Synthesis, Biomacromolecular Interactions, Photodynamic NO Releasing and Cellular Imaging of Two [RuCl(qn)(Lbpy)(NO)]X Complexes. Molecules 2021, 26, 2545. https://doi.org/10.3390/molecules26092545
Song L, Bai H, Liu C, Gong W, Wang A, Wang L, Zhao Y, Zhao X, Wang H. Synthesis, Biomacromolecular Interactions, Photodynamic NO Releasing and Cellular Imaging of Two [RuCl(qn)(Lbpy)(NO)]X Complexes. Molecules. 2021; 26(9):2545. https://doi.org/10.3390/molecules26092545
Chicago/Turabian StyleSong, Luna, Hehe Bai, Chenyang Liu, Wenjun Gong, Ai Wang, Li Wang, Yi Zhao, Xuan Zhao, and Hongfei Wang. 2021. "Synthesis, Biomacromolecular Interactions, Photodynamic NO Releasing and Cellular Imaging of Two [RuCl(qn)(Lbpy)(NO)]X Complexes" Molecules 26, no. 9: 2545. https://doi.org/10.3390/molecules26092545
APA StyleSong, L., Bai, H., Liu, C., Gong, W., Wang, A., Wang, L., Zhao, Y., Zhao, X., & Wang, H. (2021). Synthesis, Biomacromolecular Interactions, Photodynamic NO Releasing and Cellular Imaging of Two [RuCl(qn)(Lbpy)(NO)]X Complexes. Molecules, 26(9), 2545. https://doi.org/10.3390/molecules26092545