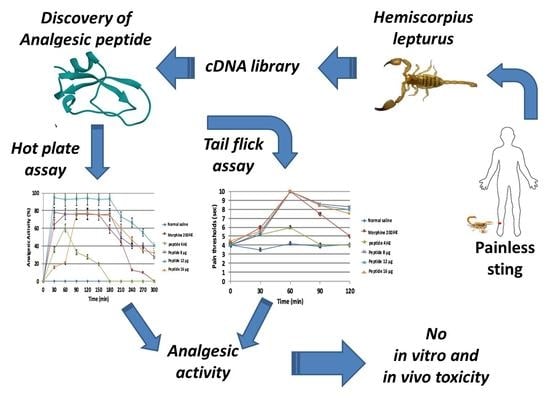
Discovery of a New Analgesic Peptide, Leptucin, from the Iranian Scorpion, Hemiscorpius lepturus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials, Cells, and Animals
2.2. Bioinformatics Analyses
2.2.1. Data Mining and ORF Determination
2.2.2. Similarity Analysis and Determination of the Mature Chain
2.2.3. Prediction of 3D Structure of the Peptide
2.3. Peptide Synthesis
2.4. Deprotection of the Synthetic Peptide
2.4.1. Quality Control of the Deprotected Peptide by SDS-PAGE
2.4.2. Evaluation of the Accuracy of the Deprotection Method by RP-HPLC
2.4.3. Determination of the Purity and Yield of the Deprotected Peptide
2.5. Refolding of the Deprotected Peptide
2.5.1. Determination of the Purity and Yield of the Refolded Peptide
2.5.2. Confirmation of Refolding by Ellman’s Assay
2.6. Dynamic Light-Scattering (DLS)
2.7. Circular Dichroism
2.8. The Evaluation of Analgesic Activity for the Refolded Leptucin
2.8.1. Hot Plate Test
2.8.2. Tail Flick Test
2.9. Toxicity Tests for Leptucin
2.9.1. MTT Test
2.9.2. In Vitro Hemolysis Assay
2.9.3. In Vivo Hemolysis Assay
2.9.4. Histopathological Study
2.9.5. Determination of Lethal Dose 50
2.10. Motor Coordination Test (Rotarod Test)
2.11. Data Analysis
3. Results
3.1. Identification of Mature Chain and Sequence Analysis
3.2. Prediction of 3D Structure of the Peptide
3.3. Deprotection Analysis
3.4. The Accuracy of Deprotection
3.5. Refolding Analysis
3.6. Refolding Confirmation
3.7. Circular Dichroism
3.8. Activity Assays
3.8.1. Hot Plate Assay
3.8.2. Tail Flick Assay
3.9. Toxicity Assays
3.10. Influence of Leptucin on Motor Coordination
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability Statement
References
- Shilpi, J.A.; Uddin, S.J. Analgesic and antipyretic natural products. Medicinal Natural Products. Dis. Focus. Approach 2020, 55, 435. [Google Scholar]
- Feng, J.; Lepetre-Mouelhi, S.; Gautier, A.; Mura, S.; Cailleau, C.; Coudore, F.; Hamon, M.; Couvreur, P. A new painkiller nanomedicine to bypass the blood-brain barrier and the use of morphine. Sci. Adv. 2019, 5, eaau5148. [Google Scholar] [CrossRef] [Green Version]
- Abd-Elsayed, A.; Deer, T.R. Different Types of Pain. In Pain; Abd-Elsayed, A., Ed.; Springer: Berlin/Heidelberg, Germany, 2019. [Google Scholar] [CrossRef]
- Hena, S.; Znad, H. Membrane bioreactor for pharmaceuticals and personal care products removal from wastewater. In Comprehensive Analyti-Cal Chemistry; Elsevier: Amsterdam, The Netherlands, 2018; Volume 81, pp. 201–256. [Google Scholar]
- Munir, M.A.; Enany, N.; Zhang, J.-M. Nonopioid Analgesics. Anesthesiol. Clin. 2007, 25, 761–774. [Google Scholar] [CrossRef]
- Kumar, M.; Shete, A.; Akbar, Z. A review on analgesic: From natural sources. Int. J. Pharm. Biol. Arch. 2010, 1, 95–100. [Google Scholar]
- Cui, J.M.; Zhao, L.; Wang, Z.J.; Ma, M.T.; Wang, Y.; Luo, K.Y.; Wang, L.Q.; Wei, S.; Zhang, X.H.; Han, C.Z.; et al. MEL endomorphins act as po-tent inflammatory analgesics with the inhibition of activated non-neuronal cells and modulation of pro-inflammatory cytokines. Neuropharmacology 2020, 168, 107992. [Google Scholar] [CrossRef]
- Bruno, B.J.; Miller, G.D.; Lim, C.S. Basics and recent advances in peptide and protein drug delivery. Ther. Deliv. 2013, 4, 1443–1467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lau, J.L.; Dunn, M.K. Therapeutic peptides: Historical perspectives, current development trends, and future directions. Bioorg. Med. Chem. 2018, 26, 2700–2707. [Google Scholar] [CrossRef]
- Maatuf, Y.; Geron, M.; Priel, A. The Role of Toxins in the Pursuit for Novel Analgesics. Toxins 2019, 11, 131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Newman, D.J.; Cragg, G.M. Natural Products as Sources of New Drugs from 1981 to 2014. J. Nat. Prod. 2016, 79, 629–661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sable, R.; Parajuli, P.; Jois, S. Peptides, Peptidomimetics, and Polypeptides from Marine Sources: A Wealth of Natural Sources for Pharmaceutical Applications. Mar. Drugs 2017, 15, 124. [Google Scholar] [CrossRef] [Green Version]
- Escoubas, P.; King, G.F. Venomics as a drug discovery platform. Expert Rev. Proteom. 2009, 6, 221–224. [Google Scholar] [CrossRef]
- Moghadasi, Z.; Shahbazzadeh, D.; Jamili, S.; Mosaffa, N.; Bagheri, K.P. Significant Anticancer Activity of a Venom Fraction Derived from the Persian Gulf Sea Anemone, Stichodactyla haddoni. Iran. J. Pharm Res. 2020, 19, 402–420. [Google Scholar] [PubMed]
- Pennington, M.W.; Czerwinski, A.; Norton, R.S. Peptide therapeutics from venom: Current status and potential. Bioorg. Med. Chem. 2018, 26, 2738–2758. [Google Scholar] [CrossRef]
- Zarghami, V.; Ghorbani, M.; Bagheri, K.P.; Shokrgozar, M.A. Melittin antimicrobial peptide thin layer on bone implant chitosan-antibiotic coatings and their bactericidal properties. Mater. Chem. Phys. 2021, 263, 124432. [Google Scholar] [CrossRef]
- Harvey, A.L. Toxins and drug discovery. Toxicon 2014, 92, 193–200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pashaei, F.; Bevalian, P.; Akbari, R.; Bagheri, K.P. Single dose eradication of extensively drug resistant Acinetobacter spp. In a mouse model of burn infection by melittin antimicrobial peptide. Microb. Pathog. 2019, 127, 60–69. [Google Scholar] [CrossRef] [PubMed]
- Khozani, R.S.; Shahbazzadeh, D.; Harzandi, N.; Feizabadi, M.M.; Bagheri, K.P. Kinetics study of antimicrobial peptide, melittin, in simultaneous bio-film degradation and eradication of potent biofilm producing MDR Pseudomonas aeruginosa isolates. Int. J. Pept. Res. Ther. 2019, 25, 329–338. [Google Scholar] [CrossRef]
- Dezfuli, H.T.; Shahbazzadeh, D.; Eidi, A.; Bagheri, K.P.; Pakravan, N.; Amini, S.; Aghasadeghi, M.R.; Mahdavi, M. Induction of IFN-γ cytokine re-sponse against hepatitis B surface antigen using melittin. Gastroenterol. Hepatol. Bed. Bench. 2014, 7, 108–117. [Google Scholar]
- Akbari, R.; Hakemi-Vala, M.; Pashaie, F.; Bevalian, P.; Hashemi, A.; Bagheri, K.P. Highly Synergistic Effects of Melittin with Conventional Antibiotics against Multidrug-Resistant Isolates of Acinetobacter baumannii and Pseudomonas aeruginosa. Microb. Drug Resist. 2019, 25, 193–202. [Google Scholar] [CrossRef] [PubMed]
- Utkin, Y.N. Animal venom studies: Current benefits and future developments. World J. Biol. Chem. 2015, 6, 28–33. [Google Scholar] [CrossRef] [PubMed]
- Ortiz, E.; Gurrola, G.B.; Schwartz, E.F.; Possani, L.D. Scorpion venom components as potential candidates for drug development. Toxicon 2015, 93, 125–135. [Google Scholar] [CrossRef]
- Kazemi, S.M.; Sabatier, J.-M. Venoms of Iranian Scorpions (Arachnida, Scorpiones) and Their Potential for Drug Discovery. Molecules 2019, 24, 2670. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chippaux, J.-P.; Goyffon, M. Epidemiology of scorpionism: A global appraisal. Acta Trop. 2008, 107, 71–79. [Google Scholar] [CrossRef] [PubMed]
- Zabihollahi, R.; Bagheri, K.P.; Keshavarz, Z.; Motevalli, F.; Bahramali, G.; Siadat, S.D.; Momen, S.B.; Shahbazzadeh, D.; Aghasadeghi, M.R. Venom Components of Iranian Scorpion Hemiscorpius lepturus Inhibit the Growth and Replication of Human Immunodeficiency Virus 1 (HIV-1). Iran. Biomed. J. 2016, 20, 259–265. [Google Scholar] [PubMed]
- Ahmadi, S.; Knerr, J.M.; Argemi, L.; Bordon, K.C.F.; Pucca, M.B.; Cerni, F.A.; Arantes, E.C.; Çalışkan, F.; Laustsen, A.H. Scorpion Venom: Detriments and Benefits. Biomedicines 2020, 8, 118. [Google Scholar] [CrossRef]
- Yang, S.; Yang, F.; Zhang, B.; Lee, B.H.; Li, B.; Luo, L.; Zheng, J.; Lai, R. A bimodal activation mechanism underlies scorpion toxin–induced pain. Sci. Adv. 2017, 3, e1700810. [Google Scholar] [CrossRef] [Green Version]
- Hakim, A.; Jiang, W.; Luo, L.; Li, B.; Yang, S.; Song, Y.; Lai, R. Scorpion Toxin, BmP01, Induces Pain by Targeting TRPV1 Channel. Toxins 2015, 7, 3671–3687. [Google Scholar] [CrossRef] [Green Version]
- Rowe, A.H.; Xiao, Y.; Scales, J.; Linse, K.D.; Rowe, M.P.; Cummins, T.R.; Zakon, H.H. Isolation and Characterization of CvIV4: A Pain Inducing α- Scorpion Toxin. PLoS ONE 2011, 6, e23520. [Google Scholar] [CrossRef]
- Torabi, E.; Behdani, M.; Chafi, M.H.; Moazzami, R.; Sabatier, J.; Khalaj, V.; Shahbazzadeh, D.; Bagheri, K.P. Characteristics and Lethality of a Novel Recombinant Dermonecrotic Venom Phospholipase D from Hemiscorpius lepturus. Toxins 2017, 9, 102. [Google Scholar] [CrossRef] [Green Version]
- Sanaei-Zadeh, H. Painless Stings of Yellow Iranian Scorpions. Iran. Red Crescent Med. J. 2017, 19, 42645. [Google Scholar] [CrossRef] [Green Version]
- Dehghani, R.; Arani, M.G. Scorpion sting prevention and treatment in ancient Iran. J. Tradit. Complement. Med. 2015, 5, 75–80. [Google Scholar] [CrossRef] [Green Version]
- Pipelzadeh, M.H.; Jalali, A.; Taraz, M.; Pourabbas, R.; Zaremirakabadi, A. An epidemiological and a clinical study on scorpionism by the Iranian scorpion Hemiscorpius lepturus. Toxicon 2007, 50, 984–992. [Google Scholar] [CrossRef] [PubMed]
- Dehghani, R.; Kamiabi, F.; Mohammadi, M. Scorpionism by Hemiscorpius spp. in Iran: A review. J. Venom. Anim. Toxins Incl. Trop. Dis. 2018, 24, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Pourkhalili, K.; Kim, E.; Mashayekhy, N.R.; Kamyab, M.; Hoseiny, S.M.; Evazy, R.; Mirakabadi, A.Z.; Seyedian, R. Cardiotoxic and Arrhythmogenic Effects of Hemiscorpius lepturus Scorpion Venom in Rats. J. Arthropod Borne Dis. 2015, 9, 215–225. [Google Scholar] [PubMed]
- Shahbazzadeh, D.; Srairi-Abid, N.; Feng, W.; Ram, N.; Borchani, L.; Ronjat, M.; Akbari, A.; Pessah, I.N.; De Waard, M.; El Ayeb, M. Hemicalcin, a new toxin from the Iranian scorpion Hemiscorpius lepturus which is active on ryanodine-sensitive Ca2+ channels. Biochem. J. 2007, 404, 89–96. [Google Scholar] [CrossRef] [Green Version]
- Jridi, I.; Catacchio, I.; Majdoub, H.; Shahbazeddah, D.; Ayeb, M.; Frassanito, M.A.; Ribatti, D.; Vacca, A.; Borchani, L. Hemilipin, a novel Hemiscor-pius lepturus venom heterodimeric phospholipase A2, which inhibits angiogenesis in vitro and in vivo. Toxicon 2015, 105, 34–44. [Google Scholar] [CrossRef] [PubMed]
- Maleki, M.; Dounighi, N.M. Purification and characterization of a novel type of neurotoxic peptides from the venom of the Iranian scorpion Hemiscorpius lepturus. Iran. J. Basic Med. Sci. 2020, 23, 195–201. [Google Scholar]
- Papadopoulos, J.S.; Agarwala, R. COBALT: Constraint-based alignment tool for multiple protein sequences. Bioinformatics 2007, 23, 1073–1079. [Google Scholar] [CrossRef] [Green Version]
- Roy, A.; Kucukural, A.; Zhang, Y. I-TASSER: A unified platform for automated protein structure and function prediction. Nat. Protoc. 2010, 5, 725–738. [Google Scholar] [CrossRef] [Green Version]
- Huang, C.C.; Meng, E.C.; Morris, J.H.; Pettersen, E.F.; Ferrin, T.E. Enhancing UCSF Chimera through web services. Nucleic Acids Res. 2014, 42, W478–W484. [Google Scholar] [CrossRef] [Green Version]
- Eisapoor, S.S.; Jamili, S.; Shahbazzadeh, D.; Mostafavi, P.G.; Bagheri, K.P. A New, High Yield, Rapid, and Cost-Effective Protocol to Deprotection of Cysteine-Rich Conopeptide, Omega-Conotoxin MVIIA. Chem. Biol. Drug Des. 2016, 87, 687–693. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Vera, E.; Walewska, A.; Olivera, A.B.M.; Bulaj, G. Role of Hydroxyprolines in the in Vitro Oxidative Folding and Biological Activity of Conotoxins. Biochemistry 2008, 47, 1741–1751. [Google Scholar] [CrossRef] [PubMed]
- Shahangian, S.S.; Sajedi, R.; Hasannia, S.; Jalili, S.; Mohammadi, M.; Taghdir, M.; Shali, A.; Mansouri, K.; Sariri, R. A conformation-based phage-display panning to screen neutralizing anti-VEGF VHHs with VEGFR2 mimicry behavior. Int. J. Biol. Macromol. 2015, 77, 222–234. [Google Scholar] [CrossRef]
- Liu, X.; Yao, G.; Wang, K.; Liu, Y.; Wan, X.; Jiang, H. Structural and Functional Characterization of Conotoxins from Conus achatinus Targeting NMDAR. Mar. Drugs 2020, 18, 135. [Google Scholar] [CrossRef] [Green Version]
- Guidance for Industry. Analgesic Indications: Developing Drug and Biological Products. Office of Communications, Division of Drug Information, Center for Drug Evaluation and Research, Food and Drug Administration. 02/06/2014. Available online: http://www.fda.gov/Drugs/GuidanceComplianceRegulatoryInformation/Guidances/default.htm (accessed on 1 December 2020).
- Moghadasi, Z.; Jamili, S.; Shahbazadeh, D.; Bagheri, K.P. Toxicity and Potential Pharmacological Activities in the Persian Gulf Venomous Sea Anemone, Stichodactyla haddoni. Iran. J. Pharm. Res. 2018, 17, 940–955. [Google Scholar]
- Carlsson, K.-H.; Jurna, I. Depression by flupirtine, a novel analgesic agent, of motor and sensory responses of the nociceptive system in the rat spinal cord. Eur. J. Pharmacol. 1987, 143, 89–99. [Google Scholar] [CrossRef]
- Regalado, A.I.; Mancebo, B.; Paixão, A.; López, Y.; Merino, N.; Sánchez, L.M. Antinociceptive Activity of Methanol Extract of Tabebuia hypoleuca (C. Wright ex Sauvalle) Urb. Stems. Med. Princ. Pract. 2017, 26, 368–374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aghazadeh, H.; Memariani, H.; Ranjbar, R.; Bagheri, K. The activity and action mechanism of novel short selective LL-37-derived anticancer peptides against clinical isolates of Escherichia coli. Chem. Biol. Drug Des. 2019, 93, 75–83. [Google Scholar] [CrossRef] [Green Version]
- Memar, B.; Jamili, S.; Shahbazzadeh, D.; Bagheri, K.P. The first report on coagulation and phospholipase A2 activities of Persian Gulf lionfish, Pterois russelli, an Iranian venomous fish. Toxicon 2016, 113, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Treuting, P.M.; Dintzis, S.M. Comparative Anatomy and Histology. In A Mouse and Human Atlas; Elsevier: London, UK, 2012; p. 461. ISBN 978-0-12-381361-9. [Google Scholar]
- Bansinath, M.; Bose, A.C.; Hema, S.; Guruswami, M.N. Interaction of metamizol with some hypnotics in rats. Arch. Int. Pharmacodyn. Ther. 1982, 260, 14–27. [Google Scholar]
- Nikolaev, M.V.; Dorofeeva, N.A.; Komarova, M.S.; Korolkova, Y.V.; Andreev, Y.A.; Mosharova, I.V.; Grishin, E.V.; Tikhonov, D.B.; Kozlov, S.A. TRPV1 activation power can switch an action mode for its polypeptide ligands. PLoS ONE 2017, 12, e0177077. [Google Scholar] [CrossRef]
- Veber, D.; Milkowski, J.; Varga, S.; Denkewalter, R.; Hirschmann, R. Acetamidomethyl. A Novel Thiol Protecting Group for Cysteine. J. Am. Chem. Soc. 1972, 94, 5456–5461. [Google Scholar] [CrossRef] [PubMed]
- Becker, S.; Atherton, E.; Gordon, R.D. Synthesis and characterization of mu-conotoxin IIIa. JBIC J. Biol. Inorg. Chem. 1989, 185, 79–84. [Google Scholar] [CrossRef]
- Steiner, A.M.; Bulaj, G. Optimization of oxidative folding methods for cysteine-rich peptides: A study of conotoxins containing three disulfide bridges. J. Pept. Sci. 2010, 17, 1–7. [Google Scholar] [CrossRef]
- O’Brien, E.P.; Dima, R.I.; Brooks, B.; Thirumalai, D. Interactions between Hydrophobic and Ionic Solutes in Aqueous Guanidinium Chloride and Urea Solutions: Lessons for Protein Denaturation Mechanism. J. Am. Chem. Soc. 2007, 129, 7346–7353. [Google Scholar] [CrossRef] [PubMed]
- Annis, I.; Hargittai, B.; Barany, G. [10] Disulfide bond formation in peptides. Branched Chain Amino Acids Part B 1997, 289, 198–221. [Google Scholar] [CrossRef]
- Buczek, P.; Buczek, O.; Bulaj, G. Total chemical synthesis and oxidative folding of delta-conotoxin PVIA containing an N-terminal propeptide. Biopolymers 2005, 80, 50–57. [Google Scholar] [CrossRef]
- Fan, X.; Zhao, F.; Wang, X.; Wu, G. Doxorubicin-triggered self-assembly of native amphiphilic peptides into spherical nanoparticles. Oncotarget 2016, 7, 58445–58458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ummenhofer, W.C.; Arends, R.H.; Shen, D.D.; Bernards, C.M. Comparative Spinal Distribution and Clearance Kinetics of Intrathecally Administered Morphine, Fentanyl, Alfentanil, and Sufentanil. Anesthesiology 2000, 92, 739–753. [Google Scholar] [CrossRef] [Green Version]










| Dissolution | pH | ||
|---|---|---|---|
| Deprotection | 4 | 7 | |
| pH | 4 | Protocol 1 | Protocol 3 |
| 7 | Protocol 2 | Protocol 4 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bagheri-Ziari, S.; Shahbazzadeh, D.; Sardari, S.; Sabatier, J.-M.; Pooshang Bagheri, K. Discovery of a New Analgesic Peptide, Leptucin, from the Iranian Scorpion, Hemiscorpius lepturus. Molecules 2021, 26, 2580. https://doi.org/10.3390/molecules26092580
Bagheri-Ziari S, Shahbazzadeh D, Sardari S, Sabatier J-M, Pooshang Bagheri K. Discovery of a New Analgesic Peptide, Leptucin, from the Iranian Scorpion, Hemiscorpius lepturus. Molecules. 2021; 26(9):2580. https://doi.org/10.3390/molecules26092580
Chicago/Turabian StyleBagheri-Ziari, Sedigheh, Delavar Shahbazzadeh, Soroush Sardari, Jean-Marc Sabatier, and Kamran Pooshang Bagheri. 2021. "Discovery of a New Analgesic Peptide, Leptucin, from the Iranian Scorpion, Hemiscorpius lepturus" Molecules 26, no. 9: 2580. https://doi.org/10.3390/molecules26092580
APA StyleBagheri-Ziari, S., Shahbazzadeh, D., Sardari, S., Sabatier, J. -M., & Pooshang Bagheri, K. (2021). Discovery of a New Analgesic Peptide, Leptucin, from the Iranian Scorpion, Hemiscorpius lepturus. Molecules, 26(9), 2580. https://doi.org/10.3390/molecules26092580