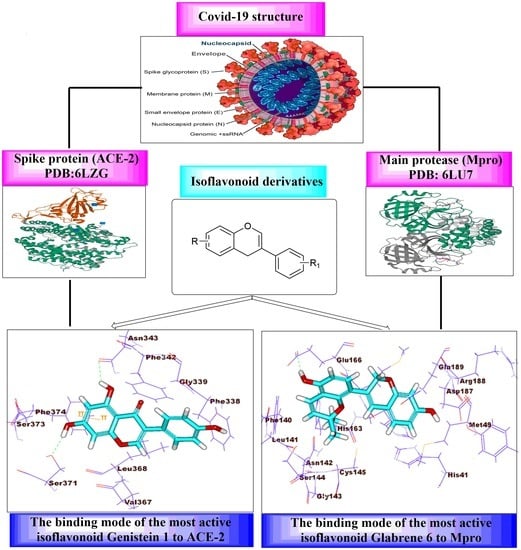
In Silico Studies of Some Isoflavonoids as Potential Candidates against COVID-19 Targeting Human ACE2 (hACE2) and Viral Main Protease (Mpro)
Abstract
:1. Introduction
2. Experimental
3. Results and Discussion
3.1. Pharmacokinetic Profiling Study
3.1.1. Lipinski’s Rule of Five
3.1.2. ADMET Studies
3.2. Molecular Docking
3.2.1. Validation Process
3.2.2. HACE2
3.2.3. Main Protease (Mpro)
3.2.4. Structure-Activity Relationship (SAR)
3.3. Toxicity Studies
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Zumla, A.; Chan, J.F.; Azhar, E.I.; Hui, D.S.; Yuen, K.Y. Coronaviruses - drug discovery and therapeutic options. Nat. Rev. Drug Discov. 2016, 15, 327–347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Su, S.; Wong, G.; Shi, W.; Liu, J.; Lai, A.C.K.; Zhou, J.; Liu, W.; Bi, Y.; Gao, G.F. Epidemiology, Genetic Recombination, and Pathogenesis of Coronaviruses. Trends Microbiol. 2016, 24, 490–502. [Google Scholar] [CrossRef] [Green Version]
- WHO. WHO Coronavirus Disease (COVID-19) Dashboard. Available online: https://covid19.who.int/ (accessed on 2 October 2020).
- Dolin, R.; Hirsch, M.S. Remdesivir—An Important First Step; Massachusetts Medical Society: Waltham, MA, USA, 2020. [Google Scholar]
- Zhang, L.; Lin, D.; Sun, X.; Curth, U.; Drosten, C.; Sauerhering, L.; Becker, S.; Rox, K.; Hilgenfeld, R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science 2020, 368, 409–412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prajapat, M.; Sarma, P.; Shekhar, N.; Avti, P.; Sinha, S.; Kaur, H.; Kumar, S.; Bhattacharyya, A.; Kumar, H.; Bansal, S. Drug targets for corona virus: A systematic review. Indian J. Pharmacol. 2020, 52, 56. [Google Scholar]
- Riordan, J.F. Angiotensin-I-converting enzyme and its relatives. Genome Biol. 2003, 4, 1–5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ichihara, K.I.; Fukubayashi, Y. Preparation of fatty acid methyl esters for gas-liquid chromatography. J. Lipid Res. 2010, 51, 635–640. [Google Scholar] [CrossRef] [Green Version]
- Ksiazek, T.G.; Erdman, D.; Goldsmith, C.S.; Zaki, S.R.; Peret, T.; Emery, S.; Tong, S.; Urbani, C.; Comer, J.A.; Lim, W. A novel coronavirus associated with severe acute respiratory syndrome. New Engl. J. Med. 2003, 348, 1953–1966. [Google Scholar] [CrossRef] [PubMed]
- Harmer, D.; Gilbert, M.; Borman, R.; Clark, K.L. Quantitative mRNA expression profiling of ACE 2, a novel homologue of angiotensin converting enzyme. Febs Lett. 2002, 532, 107–110. [Google Scholar] [CrossRef] [Green Version]
- Yan, R.; Zhang, Y.; Li, Y.; Xia, L.; Guo, Y.; Zhou, Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 2020, 367, 1444–1448. [Google Scholar] [CrossRef] [Green Version]
- Tortorici, M.A.; Veesler, D. Structural insights into coronavirus entry. In Advances in Virus Research; Elsevier: Amsterdam, The Netherlands, 2019; Volume 105, pp. 93–116. [Google Scholar]
- Yang, J.; Petitjean, S.J.; Koehler, M.; Zhang, Q.; Dumitru, A.C.; Chen, W.; Derclaye, S.; Vincent, S.P.; Soumillion, P.; Alsteens, D. Molecular interaction and inhibition of SARS-CoV-2 binding to the ACE2 receptor. Nat. Commun. 2020, 11, 1–10. [Google Scholar] [CrossRef]
- Zhang, H.; Penninger, J.M.; Li, Y.; Zhong, N.; Slutsky, A.S. Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: Molecular mechanisms and potential therapeutic target. Intensive Care Med. 2020, 46, 586–590. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jia, H.P.; Look, D.C.; Shi, L.; Hickey, M.; Pewe, L.; Netland, J.; Farzan, M.; Wohlford-Lenane, C.; Perlman, S.; McCray, P.B. ACE2 receptor expression and severe acute respiratory syndrome coronavirus infection depend on differentiation of human airway epithelia. J. Virol. 2005, 79, 14614–14621. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Metwaly, A.M.; Lianlian, Z.; Luqi, H.; Deqiang, D. Black ginseng and its saponins: Preparation, phytochemistry and pharmacological effects. Molecules 2019, 24, 1856. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.-M.; Ran, X.-K.; Riaz, M.; Yu, M.; Cai, Q.; Dou, D.-Q.; Metwaly, A.M.; Kang, T.-G.; Cai, D.-C. Chemical Constituents of Stems and Leaves of Tagetespatula L. and Its Fingerprint. Molecules 2019, 24, 3911. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sperstad, S.V.; Haug, T.; Blencke, H.-M.; Styrvold, O.B.; Li, C.; Stensvåg, K. Antimicrobial peptides from marine invertebrates: Challenges and perspectives in marine antimicrobial peptide discovery. Biotechnol. Adv. 2011, 29, 519–530. [Google Scholar] [CrossRef]
- El-Demerdash, A.; Metwaly, A.M.; Hassan, A.; El-Aziz, A.; Mohamed, T.; Elkaeed, E.B.; Eissa, I.H.; Arafa, R.K.; Stockand, J.D. Comprehensive Virtual Screening of the Antiviral Potentialities of Marine Polycyclic Guanidine Alkaloids against SARS-CoV-2 (Covid-19). Biomolecules 2021, 11, 460. [Google Scholar] [CrossRef]
- Metwaly, A.M.; Wanas, A.S.; Radwan, M.M.; Ross, S.A.; ElSohly, M.A. New α-Pyrone derivatives from the endophytic fungus Embellisia sp. Med. Chem. Res. 2017, 26, 1796–1800. [Google Scholar] [CrossRef]
- Metwaly, A.; Kadry, H.; El-Hela, A.; Elsalam, A.; Ross, S. New antimalarial benzopyran derivatives from the endophytic fungus Alternaria phragmospora. Planta Med. 2014, 80, PC11. [Google Scholar] [CrossRef]
- Metwaly, A. Comparative biological evaluation of four endophytic fungi isolated from nigella sativa seeds. Al-Azhar J. Pharm. Sci. 2019, 59, 123–136. [Google Scholar] [CrossRef]
- Yassin, A.M.; El-Deeb, N.M.; Metwaly, A.M.; El Fawal, G.F.; Radwan, M.M.; Hafez, E.E. Induction of apoptosis in human cancer cells through extrinsic and intrinsic pathways by Balanites aegyptiaca furostanol saponins and saponin-coated silvernanoparticles. Appl. Biochem. Biotechnol. 2017, 182, 1675–1693. [Google Scholar] [CrossRef]
- Sharaf, M.H.; El-Sherbiny, G.M.; Moghannem, S.A.; Abdelmonem, M.; Elsehemy, I.A.; Metwaly, A.M.; Kalaba, M.H. New combination approaches to combat methicillin-resistant Staphylococcus aureus (MRSA). Sci. Rep. 2021, 11, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Metwaly, A.M.; Ghoneim, M.M.; Musa, A. Two new antileishmanial diketopiperazine alkaloids from the endophytic fungus Trichosporum sp. Derpharmachemica 2015, 7, 322–327. [Google Scholar]
- Metwaly, A.M.; Fronczek, F.R.; Ma, G.; Kadry, H.A.; Atef, A.; Mohammad, A.-E.I.; Cutler, S.J.; Ross, S.A. Antileukemic α-pyrone derivatives from the endophytic fungus Alternaria phragmospora. Tetrahedron Lett. 2014, 55, 3478–3481. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Metwaly, A.M.; Kadry, H.A.; Atef, A.; Mohammad, A.-E.I.; Ma, G.; Cutler, S.J.; Ross, S.A. Nigrosphaerin A a new isochromene derivative from the endophytic fungus Nigrospora sphaerica. Phytochem. Lett. 2014, 7, 1–5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhanzhaxina, A.; Suleimen, Y.; Metwaly, A.M.; Eissa, I.H.; Elkaeed, E.B.; Suleimen, R.; Ishmuratova, M.; Akatan, K.; Luyten, W. In Vitro and In Silico Cytotoxic and Antibacterial Activities of a Diterpene from Cousinia alata Schrenk. J. Chem. 2021, 2021. [Google Scholar] [CrossRef]
- Ghoneim, M.M.; Afifi, W.M.; Ibrahim, M.; Elagawany, M.; Khayat, M.T.; Aboutaleb, M.H.; Metwaly, A.M. Biological evaluation and molecular docking study of metabolites from Salvadora Persica L. Growing in Egypt. Pharmacogn. Mag. 2019, 15, 232. [Google Scholar]
- Hegazy, M.M.; Metwaly, A.M.; Mostafa, A.E.; Radwan, M.M.; Mehany, A.B.M.; Ahmed, E.; Enany, S.; Magdeldin, S.; Afifi, W.M.; ElSohly, M.A. Biological and chemical evaluation of some African plants belonging to Kalanchoe species: Antitrypanosomal, cytotoxic, antitopoisomerase I activities and chemical profiling using ultra-performance liquid chromatography/quadrupole-time-of-flight mass spectrometer. Pharmacogn. Mag. 2021, 17, 6. [Google Scholar]
- Orazbekov, Y.; Datkhayev, U.; Omyrzakov, M.; Metwaly, A.; Makhatov, B.; Jacob, M.; Ramazanova, B.; Sakipova, Z.; Azembayev, A.; Orazbekuly, K. Antifungal prenylated isoflavonoids from Maclura aurantiaca. Planta Med. 2015, 81, PE10. [Google Scholar] [CrossRef]
- Dewick, P.M. Isoflavonoids. In The Flavonoids: Advances in Research since 1980, Harborne, J.B., Ed.; Springer US: Boston, MA, USA, 1988; pp. 125–209. [Google Scholar]
- Arthan, D.; Svasti, J.; Kittakoop, P.; Pittayakhachonwut, D.; Tanticharoen, M.; Thebtaranonth, Y. Antiviral isoflavonoid sulfate and steroidal glycosides from the fruits of Solanum torvum. Phytochemistry 2002, 59, 459–463. [Google Scholar] [CrossRef]
- Okubo, K.; Kudou, S.; Uchida, T.; Yoshiki, Y.; Yoshikoshi, M.; Tonomura, M. Soybean Saponin and Isoflavonoids. In Food Phytochemicals for Cancer Prevention I; American Chemical Society: Washington, DC, USA, 1993; Volume 546, pp. 330–339. [Google Scholar]
- Horio, Y.; Sogabe, R.; Shichiri, M.; Ishida, N.; Morimoto, R.; Ohshima, A.; Isegawa, Y. Induction of a 5-lipoxygenase product by daidzein is involved in the regulation of influenza virus replication. J. Clin. Biochem. Nutr. 2020, 66, 36–42. [Google Scholar] [CrossRef] [Green Version]
- Tait, S.; Salvati, A.L.; Desideri, N.; Fiore, L. Antiviral activity of substituted homoisoflavonoids on enteroviruses. Antivir. Res. 2006, 72, 252–255. [Google Scholar] [CrossRef] [PubMed]
- Desideri, N.; Olivieri, S.; Stein, M.; Sgro, R.; Orsi, N.; Conti, C. Synthesis and anti-picornavirus activity of homo-isoflavonoids. Antivir. Chem. Chemother. 1997, 8, 545–555. [Google Scholar] [CrossRef]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 1997, 23, 3–25. [Google Scholar] [CrossRef]
- BIOVIA. Discovery Studio Visualizer; BIOVIA: San Diego, CA, USA, 2012; Available online: https://discover.3ds.com/discovery-studio-visualizer-download (accessed on 22 March 2021).
- Bank. 2020. Available online: https://www.rcsb.org/structure/6LZG (accessed on 2 January 2021).
- Bank. 2020. Available online: https://www.rcsb.org/structure/6LU7 (accessed on 2 January 2020).
- Ibrahim, M.K.; Eissa, I.H.; Abdallah, A.E.; Metwaly, A.M.; Radwan, M.; ElSohly, M. Design, synthesis, molecular modeling and anti-hyperglycemic evaluation of novel quinoxaline derivatives as potential PPARγ and SUR agonists. Bioorganic Med. Chem. 2017, 25, 1496–1513. [Google Scholar] [CrossRef] [PubMed]
- El-Helby, A.G.A.; Ayyad, R.R.; Sakr, H.M.; Abdelrahim, A.S.; El-Adl, K.; Sherbiny, F.S.; Eissa, I.H.; Khalifa, M.M. Design, synthesis, molecular modeling and biological evaluation of novel 2, 3-dihydrophthalazine-1, 4-dione derivatives as potential anticonvulsant agents. J. Mol. Struct. 2017, 1130, 333–351. [Google Scholar] [CrossRef]
- Ibrahim, M.K.; Eissa, I.H.; Alesawy, M.S.; Metwaly, A.M.; Radwan, M.M.; ElSohly, M.A. Design, synthesis, molecular modeling and anti-hyperglycemic evaluation of quinazolin-4 (3H)-one derivatives as potential PPARγ and SUR agonists. Bioorganic Med. Chem. 2017, 25, 4723–4744. [Google Scholar] [CrossRef]
- Eissa, I.H.; Metwaly, A.M.; Belal, A.; Mehany, A.B.; Ayyad, R.R.; El-Adl, K.; Mahdy, H.A.; Taghour, M.S.; El-Gamal, K.M.; El-Sawah, M.E. Discovery and antiproliferative evaluation of new quinoxalines as potential DNA intercalators and topoisomerase II inhibitors. Arch. Der Pharm. 2019, 352, 1900123. [Google Scholar] [CrossRef] [PubMed]
- PerkinElmer. ChemBioDraw Ultra 14.0; PerkinElmer: Waltham, MA, USA, 2012; Available online: https://shopinformatics.perkinelmer.com/search (accessed on 2 May 2015).
- El-Gamal, K.M.; El-Morsy, A.M.; Saad, A.M.; Eissa, I.H.; Alswah, M. Synthesis, docking, QSAR, ADMET and antimicrobial evaluation of new quinoline-3-carbonitrile derivatives as potential DNA-gyrase inhibitors. J. Mol. Struct. 2018, 1166, 15–33. [Google Scholar] [CrossRef]
- El-Zahabi, M.A.; Elbendary, E.R.; Bamanie, F.H.; Radwan, M.F.; Ghareib, S.A.; Eissa, I.H. Design, synthesis, molecular modeling and anti-hyperglycemic evaluation of phthalimide-sulfonylurea hybrids as PPARγ and SUR agonists. Bioorganic Chem. 2019, 91, 103115. [Google Scholar] [CrossRef]
- Xia, X.; Maliski, E.G.; Gallant, P.; Rogers, D. Classification of kinase inhibitors using a Bayesian model. J. Med. Chem. 2004, 47, 4463–4470. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- BIOVIA. QSAR, ADMET and Predictive Toxicology. Available online: https://www.3dsbiovia.com/products/collaborative-science/biovia-discovery-studio/qsar-admet-and-predictive-toxicology.html (accessed on 15 May 2020).
- Venkatapathy, R.; Wang, N.C.Y.; Martin, T.M.; Harten, P.F.; Young, D. Structure–Activity Relationships for Carcinogenic Potential. Gen. Appl. Syst. Toxicol. 2009. [Google Scholar] [CrossRef]
- Goodrnan, G.; Wilson, R. Comparison of the dependence of the TD50 on maximum tolerated dose for mutagens and nonmutagens. Risk Anal. 1992, 12, 525–533. [Google Scholar] [CrossRef] [PubMed]
- Council, N.R. Correlation Between Carcinogenic Potency and the Maximum Tolerated Dose: Implications for Risk Assessment. In Issues in Risk Assessment; National Academies Press (US): Cambridge, MA, USA, 1993. [Google Scholar]
- Gonella Diaza, R.; Manganelli, S.; Esposito, A.; Roncaglioni, A.; Manganaro, A.; Benfenati, E. Comparison of in silico tools for evaluating rat oral acute toxicity. Sar Qsar Environ. Res. 2015, 26, 1–27. [Google Scholar] [CrossRef] [PubMed]
- Pizzo, F.; Benfenati, E. In silico models for repeated-dose toxicity (RDT): Prediction of the no observed adverse effect level (NOAEL) and lowest observed adverse effect level (LOAEL) for drugs. In In Silico Methods for Predicting Drug Toxicity; Springer: Berlin/Heidelberg, Germany, 2016; pp. 163–176. [Google Scholar]
- Venkatapathy, R.; Moudgal, C.J.; Bruce, R.M. Assessment of the oral rat chronic lowest observed adverse effect level model in TOPKAT, a QSAR software package for toxicity prediction. J. Chem. Inf. Comput. Sci. 2004, 44, 1623–1629. [Google Scholar] [CrossRef] [PubMed]
- Wilhelmus, K.R. The Draize eye test. Surv. Ophthalmol. 2001, 45, 493–515. [Google Scholar] [CrossRef]


















| Comp. | Lipinski’s Rule of Five | |||
|---|---|---|---|---|
| Log P a | Molecular Wight | HBD b | HBA c | |
| 1 | 2.14 | 270.23 | 3 | 5 |
| 2 | 2.38 | 254.23 | 2 | 4 |
| 3 | 2.36 | 284.26 | 2 | 5 |
| 4 | 3.16 | 332.34 | 2 | 6 |
| 5 | 3.17 | 302.32 | 2 | 5 |
| 6 | 3.81 | 322.35 | 2 | 4 |
| 7 | 3.09 | 328.31 | 1 | 6 |
| 8 | 2.78 | 256.25 | 3 | 4 |
| 9 | 2.68 | 270.28 | 1 | 4 |
| 10 | 3.48 | 322.35 | 1 | 4 |
| 11 | 3.23 | 338.35 | 2 | 5 |
| 12 | −1.50 | 450.41 | 1 | 10 |
| 13 | 2.12 | 300.26 | 3 | 6 |
| 14 | 2.57 | 328.31 | 1 | 6 |
| 15 | 2.10 | 330.28 | 3 | 7 |
| 16 | 2.33 | 344.31 | 2 | 7 |
| 17 | 2.55 | 358.34 | 1 | 7 |
| 18 | 2.34 | 314.28 | 2 | 6 |
| 19 | 2.60 | 326.3 | 0 | 6 |
| 20 | 2.61 | 296.27 | 0 | 5 |
| 21 | 2.36 | 284.26 | 2 | 5 |
| 22 | 2.34 | 314.28 | 2 | 6 |
| 23 | 2.58 | 356.32 | 0 | 7 |
| 24 | 2.10 | 330.28 | 3 | 7 |
| 25 | 2.36 | 284.26 | 2 | 5 |
| 26 | 2.59 | 298.2 | 1 | 5 |
| 27 | 4.84 | 358.38 | 2 | 4 |
| 28 | 6.04 | 420.49 | 2 | 5 |
| 29 | 6.07 | 420.49 | 2 | 5 |
| 30 | 5.03 | 436.49 | 3 | 6 |
| 31 | 5.03 | 436.49 | 3 | 6 |
| 32 | 3.95 | 418.43 | 2 | 6 |
| 33 | 4.78 | 420.45 | 3 | 6 |
| 34 | 3.68 | 438.4 | 4 | 7 |
| 35 | 3.73 | 436.45 | 4 | 7 |
| 36 | 2.90 | 354.35 | 4 | 6 |
| 37 | 5.61 | 422.4 | 4 | 6 |
| 38 | 3.9 | 368.38 | 3 | 6 |
| 39 | 1.91 | 288.25 | 4 | 6 |
| 40 | 2.14 | 302.27 | 3 | 6 |
| 41 | 2.46 | 316.3 | 2 | 6 |
| 42 | 2.44 | 346.33 | 2 | 7 |
| 43 | 2.465 | 316.3 | 2 | 6 |
| 44 | 2.24 | 302.27 | 3 | 6 |
| 45 | 2.48 | 332.3 | 4 | 7 |
| 46 | 1.99 | 318.278 | 4 | 7 |
| 47 | 1.88 | 302.27 | 3 | 6 |
| 48 | 4.11 | 340.37 | 3 | 5 |
| 49 | 4.09 | 370.39 | 3 | 6 |
| 50 | 4.09 | 370.39 | 3 | 6 |
| 51 | 4.32 | 384.422 | 2 | 6 |
| 52 | 4.32 | 384.42 | 2 | 6 |
| 53 | 6.19 | 422.51 | 2 | 5 |
| 54 | 6.19 | 422.51 | 2 | 5 |
| 55 | 4.32 | 384.42 | 2 | 6 |
| 56 | 5.72 | 424.48 | 4 | 6 |
| 57 | 6.21 | 392.48 | 2 | 4 |
| 58 | 4.52 | 452.49 | 3 | 7 |
| 59 | 5.95 | 438.51 | 3 | 6 |
| Compound | BBB Level a | Absorption Level b | PPB c | Solubility Level d |
|---|---|---|---|---|
| 1 | 3 | 0 | 2 | 3 |
| 2 | 2 | 0 | 2 | 3 |
| 3 | 3 | 0 | 1 | 3 |
| 4 | 2 | 0 | 0 | 3 |
| 5 | 2 | 0 | 2 | 3 |
| 6 | 1 | 0 | 0 | 2 |
| 7 | 2 | 0 | 0 | 2 |
| 8 | 2 | 0 | 1 | 3 |
| 9 | 2 | 0 | 2 | 3 |
| 10 | 1 | 0 | 2 | 2 |
| 11 | 2 | 0 | 0 | 2 |
| 12 | 4 | 2 | 0 | 4 |
| 13 | 3 | 0 | 1 | 3 |
| 14 | 3 | 0 | 2 | 3 |
| 15 | 3 | 0 | 2 | 3 |
| 16 | 3 | 0 | 1 | 3 |
| 17 | 3 | 0 | 1 | 3 |
| 18 | 3 | 0 | 2 | 3 |
| 19 | 2 | 0 | 2 | 2 |
| 20 | 2 | 0 | 1 | 2 |
| 21 | 3 | 0 | 1 | 3 |
| 22 | 3 | 0 | 1 | 3 |
| 23 | 2 | 0 | 2 | 2 |
| 24 | 3 | 0 | 2 | 3 |
| 25 | 3 | 0 | 2 | 3 |
| 26 | 2 | 0 | 2 | 3 |
| 27 | 1 | 0 | 2 | 2 |
| 28 | 4 | 1 | 2 | 2 |
| 29 | 4 | 1 | 2 | 1 |
| 30 | 4 | 1 | 2 | 2 |
| 31 | 4 | 1 | 2 | 2 |
| 32 | 2 | 0 | 0 | 2 |
| 33 | 4 | 1 | 1 | 2 |
| 34 | 4 | 1 | 0 | 2 |
| 35 | 4 | 1 | 0 | 2 |
| 36 | 4 | 0 | 1 | 3 |
| 37 | 4 | 2 | 2 | 2 |
| 38 | 4 | 0 | 0 | 2 |
| 39 | 3 | 0 | 1 | 3 |
| 40 | 3 | 0 | 1 | 3 |
| 41 | 3 | 0 | 2 | 3 |
| 42 | 3 | 0 | 2 | 3 |
| 43 | 3 | 0 | 0 | 3 |
| 44 | 3 | 0 | 2 | 3 |
| 45 | 4 | 0 | 2 | 3 |
| 46 | 4 | 0 | 0 | 3 |
| 47 | 3 | 0 | 0 | 3 |
| 48 | 2 | 0 | 1 | 2 |
| 49 | 4 | 0 | 2 | 2 |
| 50 | 4 | 0 | 1 | 2 |
| 51 | 2 | 0 | 1 | 2 |
| 52 | 2 | 0 | 2 | 2 |
| 53 | 4 | 2 | 2 | 1 |
| 54 | 4 | 2 | 2 | 1 |
| 55 | 2 | 0 | 1 | 2 |
| 56 | 4 | 2 | 2 | 2 |
| 57 | 4 | 1 | 2 | 1 |
| 58 | 4 | 1 | 1 | 2 |
| 59 | 2 | 2 | 2 | |
| Remdesivir | 4 | 3 | 0 | 2 |
| Comp. | Binding Energy (kcal mol−1) | No. of H. Bonds | Involved Amino Acid Residues | Amino Acid Residues Involved in Hydrophobic inTeraction |
|---|---|---|---|---|
| 1 | −30.90 | 2 | Ser371, Asn343 | Phe374, Gly339, Ser371, Phe338, Phe342 |
| 2 | −27.84 | 1 | Ser371 | Phe338, Phe342, Gly339 |
| 3 | −28.13 | 1 | Ser371 | Phe374, Phe342, Phe338, Gly339 |
| 4 | −25.52 | 1 | Ser371 | Phe374, Phe342, Phe338, Gly339 |
| 5 | −24.12 | 1 | Ser371 | Phe374, Phe342, Phe338, Leu368, Gly339 |
| 6 | −26.14 | 1 | Ser371 | Phe342, Phe338, Phe374 |
| 7 | −25.95 | 1 | Ser371 | Phe342, Phe338, Phe374 |
| 8 | −27.41 | 2 | Ser371, Asn343 | Phe374, Phe342, Phe338 |
| 9 | −22.32 | 1 | Ser371 | Phe374, Phe342, Phe338 |
| 10 | −23.66 | 0 | 0 | Phe374, Phe342, Phe338, Ser371, Gly339 |
| 11 | −24.02 | 1 | Ser371 | Phe374, Phe342, Phe338 |
| 12 | −31.01 | 2 | Asp364 | Phe338, Ser371, Leu368, Cys336, Phe374, Val367 |
| 13 | −27.85 | 0 | 0 | Asn343, Ser371, Leu368, Cys336, Phe374, Val367 |
| 14 | −25.17 | 1 | Cys336 | Phe338, Ser371, Ser373, Leu368, Cys336, Phe374, Val367 |
| 15 | −27.52 | 1 | Cys336 | Phe374, Phe342, Ser371, Leu368, Cys336, Val367 |
| 16 | −27.42 | 1 | Cys336 | Ser371, Leu368, Cys336, Phe374, Val367, Gly339 |
| 17 | −25.02 | 1 | Trp436 | Phe374, Leu368, Val367, Phe342 |
| 18 | −23.37 | 1 | Cys336 | Phe338, Leu368, Cys336, Phe342, Val367, Asn343 |
| 19 | −30.52 | 1 | Gly339 | Phe374, Phe338, Ser371, Gly339, Cys336, Leu368, Val367 |
| 20 | −29.50 | 0 | 0 | Phe374, Phe338, Ser371, Cys336, Leu368, Val367 |
| 21 | −24.10 | 0 | 0 | Phe338, Ser371, Cys336, Leu368, Val367, Phe374 |
| 22 | −28.66 | 1 | Cys336 | Asn434, Phe338, Ser371, Cys336, Leu368, Val367 |
| 23 | −33.20 | 0 | 0 | Phe338, Ser371, Cys336, Leu368, Val367, Ser373 |
| 24 | −32.74 | 2 | Ser371, Cys336 | Phe374, Phe338, Gly339, Cys336, Ser371, Leu368, Phe432 |
| 25 | −24.43 | 1 | Ser373 | Gly339, Leu368, Phe338, Ser371, Cys336 |
| 26 | −27.27 | 1 | Ser373 | Asn343, Gly339, Leu368, Phe338, Ser371, Cys336 |
| 27 | −30.81 | 1 | Cys336 | Phe374, Phe338, Ser371, Cys336, Leu368, Val367, Phe342, Asn343 |
| 28 | −29.91 | 0 | 0 | Leu368, Val367, Phe342, Asn343, Cys336, Phe338, Ser371, |
| 29 | −32.76 | 2 | Ser371, Cys336 | Phe374, Phe338, Ser371, Val367, Cys336, Leu368, Ser373 |
| 30 | −29.12 | 2 | Asn343, Cys336 | Phe338, Ser371, Gly339, Cys336, Leu368, Ser373, Asn343 |
| 31 | −30.84 | 1 | Asn364 | Cys336, Leu368, Ser373, Asn343, Val362, Asn364 |
| 32 | −33.95 | 0 | 0 | Phe338, Val367, Cys336, Leu368, Ser373, Asn440, Asn364 |
| 33 | −36.35 | 1 | Ser371 | Phe374, Phe342, Ser371, Asn343, Cys336, Glu340, Ser373 |
| 34 | −39.33 | 1 | Asp364 | Phe338, Phe342, Asn343, Val367, Asp364, Cys336, Leu335, Leu386 |
| 35 | −34.48 | 3 | Cys336, Gly339, Glu340 | Phe374, Phe338, Val367, Cys336, Leu368, Ser373, Gly339, Glu340 |
| 36 | −34.80 | 2 | Cys336, Gly339 | Phe338, Leu335, Asn343, Ser373 |
| 37 | −34.37 | 2 | Ser371, Ser373 | Leu368, Ser371, Asn343, Ser373, Phe338, Phe342 |
| 38 | −30.09 | 2 | Cys336, Gly339 | Phe338, Leu335, Cys336, Gly339, Asn343, Ser373 |
| 39 | −25.26 | 1 | Ser371 | Phe374, Phe338, Ser371, Val367, Cys336, Leu368, Ser373 |
| 40 | −23.32 | 1 | Ser373 | Phe338, Val367, Cys336, Leu368, Ser373 |
| 41 | −29.16 | 1 | Cys336 | Cys336, Phe338, Val367, Leu368, Ser373 |
| 42 | −32.12 | 1 | Ser371 | Phe374, Val367, Cys336, Leu368, Ser373, Phe338, Ser371, |
| 43 | −27.79 | 1 | Ser371 | Phe374, Phe338, Ser371, Val367, Cys336, Leu368 |
| 44 | −27.53 | 1 | Ser373 | Phe338, Phe374, Val367, Cys336, Leu368, Ser373 |
| 45 | −31.39 | 1 | Cys336 | Cys336, Phe342, Val367, Leu368, Gly339, Asp364 |
| 46 | −30.09 | 2 | Cys336, Gly339 | Phe338, Leu335, Cys336, Gly339, Asn343, Ser373 |
| 47 | −25.11 | 0 | 0 | Phe338, Asn343, Cys336, Leu368, Ser373 |
| 48 | −34.79 | 1 | Cys336 | Cys336, Asn343, Phe338, Val367, Leu368, Ser373 |
| 49 | −31.79 | 1 | Cys336 | Phe338, Asn343, Cys336, Leu368, Val367 |
| 50 | –30.39 | 1 | Cys336 | Cys336, Phe338, Val367, Leu368, Ser373 |
| 51 | −30.81 | 1 | Ser371 | Phe342, Asn343, Phe374, Ser371, Leu368 |
| 52 | −29.33 | 1 | Gly339 | Phe338, Leu335, Cys336, Gly339, Val367, Asn343, Ser373 |
| 53 | −33.34 | 1 | Ser373 | Phe338, Phe374, Val367, Cys336, Leu368, Ser373 |
| 54 | −35.10 | 0 | 0 | Ser371, Ser373, Phe338, Leu335, Cys336 |
| 55 | −29.06 | 1 | Cys336 | Cys336, Phe342, Val367, Leu335, Ser371, Asn343 |
| 56 | −34.90 | 2 | Ser371, Cys336 | Phe374, Phe338, Ser371, Val367, Cys336, Leu368, Ser373 |
| 57 | −34.77 | 0 | 0 | Ser373, Phe338, Phe342, Cys336, Gly339 |
| 58 | −30.22 | 1 | Ser371 | Phe342, Asn343, Phe374, Ser371, Leu368 |
| 59 | −34.70 | 1 | Ser371 | Ser373, Asn343, Phe374, Ser371, Leu368, Val367, Leu335 |
| NAG | −21.39 | 1 | Ser371. | Phe374, Phe342, Phe338 |
| Comp. | Binding Energy (kcal mol−1) | No. of H. Bonds | Involved Amino Acid Residues | Amino Acid Residues Involved in Hydrophobic Interaction |
|---|---|---|---|---|
| 1 | −37.38 | 1 | Glu166 | Phe140, Leu141, Gln189, His41, Tyr54, Glu166 |
| 2 | −35.91 | 1 | Phe140 | Phe140, Leu141, Gln189, His41, Tyr54, Glu166 |
| 3 | −36.08 | 1 | Glu166 | Phe140, Leu141, Gln189, His41, Tyr54, Glu166 |
| 4 | −37.99 | 1 | Glu166 | Phe140, Leu141, Gln189, His41, Tyr54, Glu166 |
| 5 | −38.45 | 2 | Thr190, Leu141 | Phe140, Leu141, Gln189, His41, Tyr54, Glu166 |
| 6 | −41.41 | 1 | Glu166 | Phe140, Leu141, Asn142, His163, Tyr54, Glu166 |
| 7 | −40.11 | 1 | Glu166 | Phe140, His172, Glu166, His163, His164, Gln189 |
| 8 | −42.73 | 3 | Glu166, Cys145, His163 | Phe140, Leu141, Glu166, His163, His164, Gln189 |
| 9 | −33.98 | 1 | Phe140. | Phe140, Leu141, Gln189, His41, Tyr54, Glu166 |
| 10 | −35.25 | 2 | Glu166, Phe140. | Phe140, Leu141, Gln189, His41, Tyr54, Glu166 |
| 11 | −32.19 | 1 | Glu 166 | Phe140, Leu141, Gln189, His41, Tyr54, Glu166 |
| 12 | −41.55 | 3 | Gln192, His41, Arg188 | Glu166, Met 165, Gln192, His41, His164, His172 |
| 13 | −40.51 | 1 | Glu 166 | Met 165, Cys145, His41, Asn142, Glu166 |
| 14 | −39.89 | 1 | Glu 166 | His163, Met 165, Cys145, His41, Glu189, Glu166 |
| 15 | −37.34 | 1 | Glu166 | Phe140, Met 165, Asp187, His41, Glu189, Glu166 |
| 16 | −39.05 | 6 | Glu166, Cys145, Thr26 | Glu166, Cys145, Thr26, His41, Met 165, Glu189, Leu27 |
| 17 | −40.60 | 1 | Gly143 | Glu166, Cys145, Thr26, His4, Met 165, Gln189, Gln192 |
| 18 | −35.58 | 0 | 0 | Glu166, Phe140, Gly143, Asp187, Met 165, Gln189 |
| 19 | −37.26 | 0 | 0 | Glu166, Phe140, Cys145, Asp142, Met 165, Gln189 |
| 20 | −34.97 | 1 | Glu 166 | Phe140, Gln189, His41, Ser144, Tyr54, Glu166 |
| 21 | −38.42 | 1 | Phe140 | Phe140, Leu141, Gln189, His41, Met165, Leu140, Glu166 |
| 22 | −40.14 | 1 | Phe140 | Phe140, Leu141, Gln189, His164, Met165, Leu140, Glu166 |
| 23 | −40.24 | 0 | 0 | Glu166, Phe140, Leu141, Gln189, His41, Met165, Leu140 |
| 24 | −38.90 | 2 | Phe140, Glu166 | Glu166, Phe140, Leu141, Gln189, His164, Met165, Leu140, Cys145 |
| 25 | −34.43 | 2 | Phe140, Glu166 | Glu166, Phe140, His41, Gln189, His164, Met165, Cys145 |
| 26 | −36.39 | 1 | Phe140 | Glu166, Phe140, His41, Gln189, His164, Met165 |
| 27 | −38.58 | 3 | Gly143, Cys145, Thr26 | Glu166, Gly143, Gln189, Cys145, Thr26, Met165 |
| 28 | −47.62 | 1 | His41 | Glu166, Phe140, His41, Gln189, His164, Met165, Cys145, Leu141 |
| 29 | −49.64 | 3 | Glu166, Tyr54, Asp187 | Phe140, Gln189, His172, Met165, Tyr54, Asp187, Leu167, Glu166 |
| 30 | −48.39 | 3 | Glu166, Tyr54, Asp187 | His41, Gln189, His163, Met165, Tyr54, Asp187, Leu167, Glu166 |
| 31 | −48.32 | 1 | Glu 166 | Phe140, Gln189, His41, Met165, Tyr54, Glu166 |
| 32 | −38.31 | 1 | Cys145 | Gln189, His41, Met165, Cys145, Glu166 |
| 33 | −43.52 | 2 | Gln189, Gly143 | Met165, Gln189, Gly143, Glu166 |
| 34 | −45.48 | 2 | Glu166, Cys145 | Glu166, Phe140, His41, Gln189, His164, Met165, Cys145 |
| 35 | −41.38 | 1 | Gly143 | Glu166, Gly143, Leu107, Gln192, His164, Met165, Cys145 |
| 36 | −42.29 | 4 | His164, Cys145, Ser144, Leu141 | Gln189, His172, Met165, Glu166 His164, Cys145, Ser144, Leu141 |
| 37 | −48.13 | 4 | Met165, Thr190, His41, Cys145 | Glu166, Met165, Thr190, His41, Cys145, Gln189 |
| 38 | −43.30 | 1 | Glu 166 | Gln189, His163, Met165, Ser144, Glu166, Leu167 |
| 39 | −38.05 | 4 | Glu166, Cys145 | Glu166, Cys145, Met165, Asn142 |
| 40 | −36.12 | 2 | Glu166, His163 | Glu166, His163, Phe140, Met165 |
| 41 | −38.22 | 3 | Gln189, Asp187, Tyr54 | Gln189, Met165, His163, Glu166 |
| 42 | −37.17 | 1 | Glu166 | Glu166, Leu141, Gln189, Gly143 |
| 43 | −35.41 | 1 | Asp187 | Glu166, Leu141, Met165, Ser144 |
| 44 | −36.62 | 1 | Glu166 | Gln189, Met165, His172, Glu166, His163 |
| 45 | −40.48 | 4 | Ser144, Cys145, Thr26, Gly143 | Cys145, Thr26, His163, Met165, Asn142 |
| 46 | −40.84 | 1 | His163 | Glu166, His163, Phe140, Met165 |
| 47 | −35.39 | 1 | Glu166 | Glu166, Asn142, His164, Met165 |
| 48 | −40.40 | 1 | His163 | Glu166, Leu141, Met165, Gln189 |
| 49 | −43.83 | 4 | Glu166, Cys145, His41 | Glu166, Cys145, His41, Met165, Asn142, Leu141 |
| 50 | −43.91 | 1 | Glu166 | Glu166, Leu141, Met165, Gln189 |
| 51 | −46.15 | 2 | Glu166 | Glu166, Ser144, Gln189, His41 |
| 52 | −41.20 | 1 | Glu166 | Glu166, Leu141, Met165, Gln189, Asn142 |
| 53 | −46.90 | 2 | Glu166, Phe140 | Glu166, Gln189, Leu141, Met165, His172, Phe140 |
| 54 | −50.79 | 1 | Glu166 | Glu166, Gln189, Leu141, Met165, His172 |
| 55 | −40.56 | 1 | Thr26 | Asn142, Glu166, Asn142, Leu141 |
| 56 | −48.29 | 3 | Glu166, His41 | Glu166, His41, Met165, Asn142, His164 |
| 57 | −49.89 | 2 | Gly143, Arg188 | Glu166, Gln189, Leu141, Met165, His163 |
| 58 | −42.63 | 2 | Glu166, Leu141 | Glu166, Gln189, Leu141, Met165, His172 |
| 59 | −48.11 | 2 | Gly143, Leu141 | Glu166, Gln189, Met165, |
| N3(Co-crystallized ligand) | −62.84 | 4 | Gln189, Tyr54, Asp142, Asp187. | Phe140, Glu166, His172, Thr190, Gln189, Tyr54, Asp142, Asp187. |
| Comp. | FDA Rodent Carcinogenicity | Carcinogenic Potency TD50 (Rat) a | Rat MTD (Feed) b | Rat Oral LD50 c | Rat Chronic LOAEL d | Ocular Irritancy | Skin Irritancy |
|---|---|---|---|---|---|---|---|
| 1 | Non-Carcinogen | 60.47 | 0.516 | 1.40 | 0.107 | Irritant | None |
| 2 | Non-Carcinogen | 67.14 | 0.334 | 1.41 | 0.089 | Irritant | None |
| 3 | Non-Carcinogen | 10.43 | 0.225 | 0.81 | 0.068 | Irritant | None |
| 4 | Non-Carcinogen | 5.69 | 0.231 | 0.17 | 0.072 | Irritant | None |
| 5 | Non-Carcinogen | 5.73 | 0.234 | 0.17 | 0.071 | Irritant | None |
| 6 | Carcinogen | 35.33 | 0.239 | 0.77 | 0.024 | Irritant | None |
| 7 | Non-Carcinogen | 6.23 | 0.096 | 0.48 | 0.019 | Irritant | None |
| 8 | Non-Carcinogen | 33.45 | 0.529 | 1.06 | 0.074 | Irritant | Mild |
| 9 | Carcinogen | 4.43 | 0.122 | 0.14 | 0.027 | Irritant | Mild |
| 10 | Carcinogen | 28.52 | 0.126 | 0.16 | 0.011 | Irritant | None |
| 11 | Non-Carcinogen | 7.51 | 0.192 | 0.55 | 0.015 | Irritant | None |
| 12 | Non-Carcinogen | 193.96 | 0.078 | 0.10 | 0.004 | Mild | None |
| 13 | Non-Carcinogen | 5.27 | 0.255 | 1.07 | 0.865 | Mild | None |
| 14 | Non-Carcinogen | 9.10 | 0.164 | 1.13 | 0.325 | Mild | None |
| 15 | Non-Carcinogen | 7.32 | 0.288 | 2.03 | 0.147 | Mild | None |
| 16 | Non-Carcinogen | 7.91 | 0.230 | 1.67 | 0.155 | Mild | None |
| 17 | Non-Carcinogen | 8.98 | 0.184 | 1.18 | 0.152 | None | None |
| 18 | Non-Carcinogen | 8.40 | 0.205 | 1.69 | 0.309 | Mild | None |
| 19 | Non-Carcinogen | 0.77 | 0.069 | 0.39 | 0.130 | None | Mild |
| 20 | Non-Carcinogen | 0.59 | 0.061 | 0.20 | 0.145 | None | Mild |
| 21 | Non-Carcinogen | 6.40 | 0.181 | 1.44 | 0.229 | Mild | None |
| 22 | Non-Carcinogen | 5.73 | 0.205 | 2.36 | 0.390 | Mild | None |
| 23 | Non-Carcinogen | 0.44 | 0.077 | 0.42 | 0.282 | Mild | Mild |
| 24 | Non-Carcinogen | 6.88 | 0.288 | 4.66 | 0.863 | Mild | None |
| 25 | Non-Carcinogen | 19.50 | 0.181 | 0.97 | 0.191 | None | None |
| 26 | Non-Carcinogen | 10.75 | 0.145 | 1.01 | 0.281 | Mild | None |
| 27 | Non-Carcinogen | 29.81 | 0.184 | 1.74 | 0.054 | Mild | None |
| 28 | Non-Carcinogen | 19.03 | 0.199 | 0.77 | 0.035 | None | None |
| 29 | Non-Carcinogen | 25.03 | 0.080 | 0.35 | 0.055 | Severe | None |
| 30 | Non-Carcinogen | 2.33 | 0.097 | 0.45 | 0.039 | Severe | None |
| 31 | Non-Carcinogen | 2.33 | 0.097 | 0.45 | 0.039 | Severe | None |
| 32 | Non-Carcinogen | 20.46 | 0.128 | 0.26 | 0.074 | Mild | None |
| 33 | Non-Carcinogen | 73.66 | 0.197 | 0.29 | 0.013 | Mild | Mild |
| 34 | Non-Carcinogen | 25.44 | 0.526 | 0.92 | 0.018 | Severe | None |
| 35 | Non-Carcinogen | 6.87 | 0.236 | 0.37 | 0.013 | Mild | None |
| 36 | Non-Carcinogen | 322.42 | 0.764 | 0.84 | 0.029 | Severe | None |
| 37 | Non-Carcinogen | 165.35 | 0.303 | 1.39 | 0.008 | Mild | None |
| 38 | Non-Carcinogen | 19.21 | 0.153 | 0.46 | 0.024 | Mild | None |
| 39 | Non-Carcinogen | 35.43 | 0.576 | 0.70 | 0.012 | Severe | None |
| 40 | Non-Carcinogen | 4.926 | 0.216 | 0.44 | 0.015 | Mild | None |
| 41 | Non-Carcinogen | 6.31 | 0.381 | 0.98 | 0.075 | Severe | None |
| 42 | Non-Carcinogen | 5.95 | 0.428 | 0.71 | 0.026 | Mild | None |
| 43 | Non-Carcinogen | 6.31 | 0.381 | 0.91 | 0.044 | Mild | None |
| 44 | Non-Carcinogen | 5.81 | 0.475 | 1.12 | 0.041 | Mild | None |
| 45 | Non-Carcinogen | 5.28 | 0.402 | 0.76 | 0.037 | Mild | None |
| 46 | Non-Carcinogen | 3.25 | 0.668 | 1.10 | 0.174 | Mild | None |
| 47 | Non-Carcinogen | 4.02 | 0.395 | 0.65 | 0.084 | None | None |
| 48 | Non-Carcinogen | 126.90 | 0.545 | 0.39 | 0.009 | Severe | None |
| 49 | Non-Carcinogen | 14.44 | 0.284 | 0.32 | 0.024 | Mild | None |
| 50 | Non-Carcinogen | 14.44 | 0.284 | 0.20 | 0.008 | Mild | None |
| 51 | Non-Carcinogen | 16.34 | 0.226 | 0.14 | 0.008 | Mild | None |
| 52 | Non-Carcinogen | 21.43 | 0.226 | 0.46 | 0.010 | Mild | None |
| 53 | Non-Carcinogen | 18.79 | 0.150 | 0.34 | 0.008 | Mild | None |
| 54 | Non-Carcinogen | 18.79 | 0.150 | 0.26 | 0.007 | Mild | None |
| 55 | Non-Carcinogen | 14.61 | 0.226 | 0.32 | 0.053 | Severe | None |
| 56 | Non-Carcinogen | 116.75 | 0.562 | 0.42 | 0.006 | Severe | None |
| 57 | Non-Carcinogen | 177.62 | 0.291 | 0.36 | 0.004 | Severe | None |
| 58 | Non-Carcinogen | 5.28 | 0.156 | 0.18 | 0.016 | Mild | None |
| 59 | Non-Carcinogen | 15.21 | 0.208 | 0.35 | 0.014 | Mild | None |
| Simeprevir | Non-Carcinogen | 0.28 | 0.003 | 0.21 | 0.002 | Irritant | None |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alesawy, M.S.; Abdallah, A.E.; Taghour, M.S.; Elkaeed, E.B.; H. Eissa, I.; Metwaly, A.M. In Silico Studies of Some Isoflavonoids as Potential Candidates against COVID-19 Targeting Human ACE2 (hACE2) and Viral Main Protease (Mpro). Molecules 2021, 26, 2806. https://doi.org/10.3390/molecules26092806
Alesawy MS, Abdallah AE, Taghour MS, Elkaeed EB, H. Eissa I, Metwaly AM. In Silico Studies of Some Isoflavonoids as Potential Candidates against COVID-19 Targeting Human ACE2 (hACE2) and Viral Main Protease (Mpro). Molecules. 2021; 26(9):2806. https://doi.org/10.3390/molecules26092806
Chicago/Turabian StyleAlesawy, Mohamed S., Abdallah E. Abdallah, Mohammed S. Taghour, Eslam B. Elkaeed, Ibrahim H. Eissa, and Ahmed M. Metwaly. 2021. "In Silico Studies of Some Isoflavonoids as Potential Candidates against COVID-19 Targeting Human ACE2 (hACE2) and Viral Main Protease (Mpro)" Molecules 26, no. 9: 2806. https://doi.org/10.3390/molecules26092806
APA StyleAlesawy, M. S., Abdallah, A. E., Taghour, M. S., Elkaeed, E. B., H. Eissa, I., & Metwaly, A. M. (2021). In Silico Studies of Some Isoflavonoids as Potential Candidates against COVID-19 Targeting Human ACE2 (hACE2) and Viral Main Protease (Mpro). Molecules, 26(9), 2806. https://doi.org/10.3390/molecules26092806