Ligation Motifs in Zinc-Bound Sulfonamide Drugs Assayed by IR Ion Spectroscopy
Abstract
:1. Introduction
2. Results and Discussion
2.1. Mass Spectrometry and Photofragmentation Patterns
2.2. Structural and Vibrational Features of Deprotonated Sulfadiazine, (SDZ−H)−, Ions
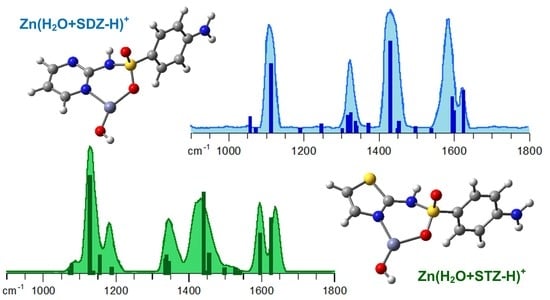
2.3. Structural and Vibrational Features of Zn(H2O+SDZ−H)+ Complexes
2.4. Structural and Vibrational Features of Zn(H2O+STZ−H)+ Complexes
3. Concluding Remarks: Ligation Motifs in SDZ- and STZ-Coordinated Zinc Complexes Assayed as Isolated Species in the Gas Phase
4. Materials and Methods
4.1. Sample Solutions for Electrospray Ionization
4.2. IRMPD Spectroscopy
4.3. Computational Methods
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Christensen, S.B. Drugs that changed society: History and current status of the early antibiotics: Salvarsan, sulfonamides, and β-lactams. Molecules 2021, 26, 6057. [Google Scholar] [CrossRef]
- Azevedo-Barbosa, H.; Dias, D.F.; Franco, L.L.; Hawkes, J.A.; Carvalho, D.T. From antibacterial to antitumour agents: A brief review on the chemical and medicinal aspects of sulfonamides. Mini-Rev. Med. Chem. 2020, 20, 2052–2066. [Google Scholar] [CrossRef]
- Ovung, A.; Bhattacharyya, J. Sulfonamide drugs: Structure, antibacterial property, toxicity, and biophysical interactions. Biophys. Rev. 2021, 13, 259–272. [Google Scholar] [CrossRef]
- Supuran, C.T. Special issue: Sulfonamides. Molecules 2017, 22, 1642. [Google Scholar] [CrossRef] [Green Version]
- Duan, W.; Cui, H.; Jia, X.; Huang, X. Occurrence and ecotoxicity of sulfonamides in the aquatic environment: A review. Sci. Total Environ. 2022, 820, 153178. [Google Scholar] [CrossRef]
- Sukul, P.; Spiteller, M. Sulfonamides in the environment as veterinary drugs. Rev. Environ. Contam. Toxicol. 2006, 187, 67–101. [Google Scholar]
- Supuran, C.T.; Minicione, F.; Scozzafava, A.; Briganti, F.; Mincione, G.; Ilises, M.A. Metal complexes of heterocyclic sulfonamides: A new class of strong topical intraocular pressure-lowering agents in rabbits. Eur. J. Med. Chem. 1998, 33, 247–254. [Google Scholar] [CrossRef]
- Pervaiz, M.; Riaz, A.; Munir, A.; Saeed, Z.; Hussain, S.; Rashid, A.; Younas, U.; Adnan, A. Synthesis and characterization of sulfonamide metal complexes as antimicrobial agents. J. Mol. Struct. 2020, 1202, 127284. [Google Scholar] [CrossRef]
- Mastrolorenzo, A.; Scozzafava, A.; Supuran, C.T. Antifungal activity of silver and zinc complexes of sulfadrug derivatives incorporating arylsulfonylureido moieties. Eur. J. Pharm. Sci. 2000, 11, 99–107. [Google Scholar] [CrossRef]
- Fox, C.L.; Rao, T.N.; Azmeth, R.; Gandhi, S.S.; Modak, S. Comparative evaluation of zinc sulfadiazine and silver sulfadiazine in burn wound infection. J. Burn Care Rehabil. 1990, 11, 112–117. [Google Scholar] [CrossRef]
- Nocentini, A.; Donald, W.A.; Supuran, C.T. Chapter 8—Human carbonic anhydrases tissue distribution, physiological role, and druggability. In Carbonic Anhydrases; Nocentini, A., Supuran, C.T., Eds.; Elsevier: Amsterdam, The Netherlands, 2019; pp. 151–185. [Google Scholar]
- Carta, F.; Supuran, C.T.; Scozzafava, A. Sulfonamides and their isosters as carbonic anhydrase inhibitors. Future Med. Chem. 2014, 6, 1149–1165. [Google Scholar] [CrossRef]
- D’Ascenzio, M.; Secci, D.; Carradori, S.; Zara, S.; Guglielmi, P.; Cirilli, R.; Pierini, M.; Poli, G.; Tuccinardi, T.; Angeli, A.; et al. 1,3-Dipolar Cycloaddition, HPLC Enantioseparation, and Docking Studies of Saccharin/Isoxazole and Saccharin/Isoxazoline Derivatives as Selective Carbonic Anhydrase IX and XII Inhibitors. J. Med. Chem. 2020, 63, 2470–2488. [Google Scholar] [CrossRef]
- Zoppi, C.; Nocentini, A.; Supuran, C.T.; Pratesi, A.; Messori, L. Native mass spectrometry of human carbonic anhydrase I and its inhibitor complexes. J. Biol. Inorg. Chem. 2020, 25, 979–993. [Google Scholar] [CrossRef]
- Uhlemann, T.; Berden, G.; Oomens, J. Preferred protonation site of a series of sulfa drugs in the gas phase revealed by IR spectroscopy. Eur. Phys. J. D 2021, 75, 23. [Google Scholar] [CrossRef]
- Chourasiya, S.S.; Patel, D.R.; Nagaraja, C.M.; Chakraborti, A.K.; Bharatam, P.V. Sulfonamide vs. sulfonimide: Tautomerism and electronic structure analysis of N-heterocyclic arenesulfonamides. New J. Chem. 2017, 41, 8118–8129. [Google Scholar] [CrossRef]
- Polfer, N.C.; Oomens, J. Vibrational spectroscopy of bare and solvated ionic complexes of biological relevance. Mass Spectrom. Rev. 2009, 28, 468–494. [Google Scholar] [CrossRef]
- Eyler, J.R. Infrared multiple photon dissociation spectroscopy of ions in Penning traps. Mass Spectrom. Rev. 2009, 28, 448–467. [Google Scholar] [CrossRef]
- Fridgen, T.D. Infrared consequence spectroscopy of gaseous protonated and metal ion cationized complexes. Mass Spectrom. Rev. 2009, 28, 586–607. [Google Scholar] [CrossRef]
- Stedwell, C.N.; Galindo, J.F.; Roitberg, A.E.; Polfer, N.C. Structures of biomolecular ions in the gas phase probed by infrared light sources. Annu. Rev. Anal. Chem. 2013, 6, 267–285. [Google Scholar] [CrossRef]
- Jašíková, L.; Roithová, J. Infrared multiphoton dissociation spectroscopy with free-electron lasers: On the road from small molecules to biomolecules. Chem. Eur. J. 2018, 24, 3374–3390. [Google Scholar] [CrossRef] [Green Version]
- Nieto, P.; Günther, A.; Berden, G.; Oomens, J.; Dopfer, O. IRMPD spectroscopy of metalated flavins: Structure and bonding of lumiflavin complexes with alkali and coinage metal ions. J. Phys. Chem. A 2016, 120, 8297–8308. [Google Scholar] [CrossRef]
- Dunbar, R.C.; Martens, J.; Berden, G.; Oomens, J. Transition metal(II) complexes of histidine-containing tripeptides: Structures, and infrared spectroscopy by IRMPD. Int. J. Mass Spectrom. 2018, 429, 198–205. [Google Scholar] [CrossRef]
- Berdakin, M.; Steinmetz, V.; Maitre, P.; Pino, G.A. On the Ag+-cytosine interaction: The effect of microhydration probed by IR optical spectroscopy and density functional theory. Phys. Chem. Chem. Phys. 2015, 17, 25915–25924. [Google Scholar] [CrossRef]
- He, C.C.; Hamlow, L.A.; Kimutai, B.; Roy, H.A.; Devereaux, Z.J.; Cunningham, N.A.; Martens, J.; Berden, G.; Oomens, J.; Chow, C.S.; et al. Structural determination of arginine-linked cisplatin complexes via IRMPD action spectroscopy: Arginine binds to platinum via NO− binding mode. Phys. Chem. Chem. Phys. 2021, 23, 21959–21971. [Google Scholar] [CrossRef]
- Stevenson, B.C.; Peckelsen, K.; Martens, J.; Berden, G.; Oomens, J.; Schäfer, M.; Armentrout, P.B. An investigation of inter-ligand coordination and flexibility: IRMPD spectroscopic and theoretical evaluation of calcium and nickel histidine dimers. J. Mol. Spectrosc. 2021, 381, 111532. [Google Scholar] [CrossRef]
- Corinti, D.; Crestoni, M.E.; Chiavarino, B.; Fornarini, S.; Scuderi, D.; Salpin, J.-Y. Insights into Cisplatin Binding to Uracil and Thiouracils from IRMPD Spectroscopy and Tandem Mass Spectrometry. J. Am. Soc. Mass Spectrom. 2020, 31, 946–960. [Google Scholar] [CrossRef]
- Corinti, D.; Coletti, C.; Re, N.; Paciotti, R.; Maitre, P.; Chiavarino, B.; Crestoni, M.E.; Fornarini, S. Short-lived intermediates (encounter complexes) in cisplatin ligand exchange elucidated by infrared ion spectroscopy. Int. J. Mass Spectrom. 2019, 435, 7–17. [Google Scholar] [CrossRef]
- Paciotti, R.; Corinti, D.; Maitre, P.; Coletti, C.; Re, N.; Chiavarino, B.; Crestoni, M.E.; Fornarini, S. From preassociation to chelation: A survey of cisplatin interaction with methionine at molecular level by IR ion spectroscopy and computations. J. Am. Soc. Mass Spectrom. 2021, 32, 2206–2217. [Google Scholar] [CrossRef]
- Corinti, D.; Crestoni, M.E.; Fornarini, S.; Dabbish, E.; Sicilia, E.; Gabano, E.; Perin, E.; Osella, D. A multi-methodological inquiry of the behavior of cisplatin-based Pt(IV) derivatives in the presence of bioreductants with a focus on the isolated encounter complexes. J. Biol. Inorg. Chem. 2020, 25, 655–670. [Google Scholar] [CrossRef]
- Corinti, D.; Maccelli, A.; Chiavarino, B.; Maitre, P.; Scuderi, D.; Bodo, E.; Fornarini, S.; Crestoni, M.E. Vibrational signatures of curcumin’s chelation in copper(II) complexes: An appraisal by IRMPD spectroscopy. J. Chem. Phys. 2019, 150, 165101. [Google Scholar] [CrossRef]
- Corinti, D.; Paciotti, R.; Re, N.; Coletti, C.; Chiavarino, B.; Crestoni, M.E.; Fornarini, S. Binding motifs of cisplatin interaction with simple biomolecules and aminoacid targets probed by IR ion spectroscopy. Pure Appl. Chem. 2020, 92, 3–13. [Google Scholar] [CrossRef]
- Boles, G.C.; Stevenson, B.C.; Hightower, R.L.; Berden, G.; Oomens, J.; Armentrout, P.B. Zinc and cadmium complexation of L-methionine: An infrared multiple photon dissociation spectroscopy and theoretical study. J. Mass Spectrom. 2021, 56, e4580. [Google Scholar] [CrossRef]
- Stevenson, B.C.; Martens, J.; Berden, G.; Oomens, J.; Schäfer, M.; Armentrout, P.B. IRMPD spectroscopic and theoretical structural investigations of zinc and cadmium dications bound to histidine dimers. J. Phys. Chem. A 2020, 124, 10266–10276. [Google Scholar] [CrossRef]
- Polfer, N.C.; Oomens, J.; Moore, D.T.; von Helden, G.; Meijer, G.; Dunbar, R.C. Infrared spectroscopy of phenylalanine Ag(I) and Zn(II) complexes in the gas phase. J. Am. Chem. Soc. 2006, 128, 517–525. [Google Scholar] [CrossRef]
- Power, B.; Haldys, V.; Salpin, J.-Y.; Fridgen, T.D. Structures of [M(Ura-H)(Ura)]+ and [M(Ura-H)(H2O)n]+ (M = Cu, Zn, Pb; n = 1–3) complexes in the gas phase by IRMPD spectroscopy in the fingerprint region and theoretical studies. Int. J. Mass Spectrom. 2018, 429, 56–65. [Google Scholar] [CrossRef] [Green Version]
- Gholami, A.; Fridgen, T.D. Structures and unimolecular reactivity of gas-phase [Zn(Proline-H)]+ and [Zn(Proline-H)(H2O)]+. J. Phys. Chem. B 2013, 117, 8447–8456. [Google Scholar] [CrossRef]
- Lagutschenkov, A.; Lorenz, U.J.; Dopfer, O. IR spectroscopy of isolated metal-organic complexes of biocatalytic interest: Evidence for coordination number four for Zn2+(imidazole)4. Int. J. Mass Spectrom. 2011, 308, 316–329. [Google Scholar] [CrossRef]
- Oomens, J.; Sartakov, B.G.; Meijer, G.; von Helden, G. Gas-phase infrared multiple photon dissociation spectroscopy of mass-selected molecular ions. Int. J. Mass Spectrom. 2006, 254, 1–19. [Google Scholar] [CrossRef]
- MacAleese, L.; Maitre, P. Infrared spectroscopy of organometallic ions in the gas phase: From model to real world complexes. Mass Spectrom. Rev. 2007, 26, 583–605. [Google Scholar] [CrossRef]
- Fornarini, S. Mass spectrometry of sulfonic acids and their derivatives. In The Chemistry of Sulphonic Acids, Esters and Their Derivatives; Patai, S., Rappoport, Z., Eds.; Wiley: Chichester, UK, 1991; pp. 73–133. [Google Scholar]
- Hu, N.; Liu, P.; Jiang, K.; Zhou, Y.; Pan, Y. Mechanism study of SO2 elimination from sulfonamides by negative electrospray ionization mass spectrometry. Rapid Comm. Mass Spectrom. 2008, 22, 2715–2722. [Google Scholar] [CrossRef]
- Corinti, D.; Crestoni, M.E.; Fornarini, S.; Pieper, M.; Niehaus, K.; Giampà, M. An integrated approach to study novel properties of a MALDI matrix (4-maleicanhydridoproton sponge) for MS imaging analyses. Anal. Bioanal. Chem. 2019, 411, 953–964. [Google Scholar] [CrossRef] [Green Version]
- Henion, J.D.; Thomson, B.A.; Dawson, P.H. Determination of Sulfa Drugs in Biological Fluids by Liquid Chromatography/Mass Spectrometry/Mass Spectrometry. Anal. Chem. 1982, 54, 451–456. [Google Scholar] [CrossRef] [PubMed]
- Klagkou, K.; Pullen, F.; Harrison, M.; Organ, A.; Firth, A.; Langley, G.J. Fragmentation pathways of sulphonamides under electrospray tandem mass spectrometric conditions. Rapid Commun. Mass Spectrom. 2003, 17, 2373–2379. [Google Scholar] [CrossRef] [PubMed]
- Guo, N.; Shen, S.; Song, W.; Pan, Y. Intramolecular oxygen transfer in the gas-phase dissociation of protonated sulfonamides. Int. J. Mass Spectrom. 2019, 435, 124–128. [Google Scholar] [CrossRef]
- Wang, S.; Guo, C.; Zhang, N.; Wu, Y.; Zhang, H.; Jiang, K. Tosyl oxygen transfer and ion-neutral complex mediated electron transfer in the gas-phase fragmentation of the protonated N-phenyl p-toluenesulfonamides. Int. J. Mass Spectrom. 2015, 376, 6–12. [Google Scholar] [CrossRef]
- Barry, S.J.; Wolff, J.-C. Identification of isobaric amino-sulfonamides without prior separation. Rapid. Commun. Mass Spectrom. 2012, 26, 419–429. [Google Scholar]
- Liu, K.; Xu, S.; Zhang, M.; Kou, Y.; Zhou, X.; Luo, K.; Hu, L.; Liu, X.; Liu, M.; Bai, L. Estimation of the toxicity of sulfadiazine to Daphnia magna using negligible depletion hollowfiber liquid-phase microextraction independent of ambient pH. Sci. Rep. 2016, 6, 39798. [Google Scholar] [CrossRef]
- Ogruc-Ildiz, G.; Akyuz, S.; Ozel, A.E. Experimental, ab initio and density functional theory studies on sulfadiazine. J. Mol. Struct. 2009, 924, 514–522. [Google Scholar] [CrossRef]
- Casanova, J.; Alzuet, G.; Ferrer, S.; Borras, J.; García-Granda, S.; Perez-Carreno, E. Metal complexes of sulfanilamide derivatives. Crystal structure of [Zn(sulfathiazole)2]·H2O. J. Inorg. Biochem. 1993, 51, 689–699. [Google Scholar] [CrossRef]
- Marsh, B.M.; Voss, J.M.; Zhou, J.; Garand, E. Coordination structure and charge transfer in microsolvated transition metal hydroxide clusters [MOH]+(H2O)1–4. Phys. Chem. Chem. Phys. 2015, 17, 23195–23206. [Google Scholar] [CrossRef] [Green Version]
- Berg, J.M.; Shi, Y. The galvanization of biology: A growing appreciation for the roles of zinc. Science 1996, 271, 1081–1085. [Google Scholar] [CrossRef] [PubMed]
- Cooper, T.E.; Armentrout, P.B. Experimental and theoretical investigation of the charge-separation energies of hydrated zinc(II): Redefinition of the critical size. J. Phys. Chem. A 2009, 113, 13742–13751. [Google Scholar] [CrossRef] [PubMed]
- Herber, I.; Tang, W.-K.; Wong, H.-Y.; Lam, T.-W.; Siu, C.-K.; Beyer, M.K. Reactivity of hydrated monovalent first row transition metal ions [M(H2O)n]+, M = Cr, Mn, Fe, Co, Ni, Cu, and Zn, n < 50, toward acetonitrile. J. Phys. Chem. A 2015, 119, 5566–5578. [Google Scholar] [PubMed]
- Bakker, J.M.; Besson, T.; Lemaire, J.; Scuderi, D.; Maitre, P. Gas-phase structure of a π-allyl palladium complex: Efficient infrared spectroscopy in a 7 T Fourier transform mass spectrometer. J. Phys. Chem. A 2007, 111, 13415–13424. [Google Scholar] [CrossRef] [PubMed]
- Corinti, D.; Maccelli, A.; Crestoni, M.E.; Cesa, S.; Quaglio, D.; Botta, B.; Ingallina, C.; Mannina, L.; Tintaru, A.; Chiavarino, B.; et al. IR ion spectroscopy in a combined approach with MS/MS and IM-MS to discriminate epimeric anthocyanin glycosides (cyanidin 3-O-glucoside and -galactoside). Int. J. Mass Spectrom. 2019, 444, 116179. [Google Scholar] [CrossRef]
- Wavefuntion. Spartan 16; Wavefuntion Inc.: Irvine, CA, USA, 2016. [Google Scholar]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A.; et al. Gaussian 09, Revision D.01; Gaussian, Inc.: Wallingford, CT, USA, 2010. [Google Scholar]
- Paciotti, R.; Coletti, C.; Re, N.; Scuderi, D.; Chiavarino, B.; Fornarini, S.; Crestoni, M.E. Serine O-sulfation probed by IRMPD spectroscopy. Phys. Chem. Chem. Phys. 2015, 17, 25891–25904. [Google Scholar] [CrossRef]
- Correia, C.F.; Balaj, P.O.; Scuderi, D.; Maitre, P.; Ohanessian, G. Vibrational signatures of protonated, phosphorylated amino acids in the gas phase. J. Am. Chem. Soc. 2008, 130, 3359–3370. [Google Scholar] [CrossRef]
- Scuderi, D.; Correia, C.F.; Balaj, O.P.; Ohanessian, G.; Lemaire, J.; Maitre, P. Structural characterization by IRMPD spectroscopy and DFT calculations of deprotonated phosphorylated amino acids in the gas phase. ChemPhysChem 2009, 10, 1630–1641. [Google Scholar] [CrossRef]
- Sinha, R.K.; Chiavarino, B.; Fornarini, S.; Lemaire, J.; Maitre, P.; Crestoni, M.E. Protonated sulfuric acid: Vibrational signatures of the naked ion in the near- and Mid-IR. J. Phys. Chem. Lett. 2010, 1, 1721–1724. [Google Scholar] [CrossRef]
- Nei, Y.-W.; Hallowita, N.; Steill, J.D.; Oomens, J.; Rodgers, M.T. Infrared multiple photon dissociation action spectroscopy of deprotonated DNA mononucleotides: Gas-phase conformations and energetics. J. Phys. Chem. A 2013, 117, 1319–1335. [Google Scholar] [CrossRef]
- O’Brien, J.T.; Prell, J.S.; Berden, G.; Oomens, J.; Williams, E.R. Effects of anions on the zwitterion stability of Glu, His and Arg investigated by IRMPD spectroscopy and theory. Int. J. Mass Spectrom. 2010, 297, 116–123. [Google Scholar] [CrossRef]
- Chiavarino, B.; Sinha, R.K.; Crestoni, M.E.; Corinti, D.; Filippi, A.; Fraschetti, C.; Scuderi, D.; Maitre, P.; Fornarini, S. Binding Motifs in the Naked Complexes of Target Amino Acids with an Excerpt of Antitumor Active Biomolecule: An Ion Vibrational Spectroscopy Assay. Chem. Eur. J. 2021, 27, 2348–2360. [Google Scholar] [CrossRef] [PubMed]




| IRMPD 1 | Calculated SDZ−H_1 1,2 | Assignment |
|---|---|---|
| 1579 | 1608 (101) | NH2 sciss |
| 1580 (351) | CN (pyr) stretch | |
| 1517 | 1505 (96) | CC (pyr) stretch |
| 1456 | 1442 (1291) | C(pyr)-NSO2 stretch |
| 1289 | 1282 (182) | SO2 asymm stretch |
| 1156 | 1144 (347) | SO2 symm stretch |
| 1103 | 1100 (135) | C-S stretch |
| IRMPD 1 | Calculated OHD_1 1,2 | Assignment |
|---|---|---|
| 1622 | 1624 (356) | NH2 sciss |
| 1582 | 1601 (172) | C-C(pyr)+C-N(pyr) stretch |
| 1601 (149) | C-C(pyr)+C-N(pyr) stretch | |
| 1594 (298) | C-C(aniline) stretch | |
| 1430 | 1429 (784) | C(pyr)-NSO2 stretch |
| 1322 | 1325 (159) | SO2 asymm stretch |
| 1318 (132) | C(pyr)-NSO2 stretch + CH bend | |
| 1108 | 1111 (582) | C-S stretch + S-O stretch |
| IRMPD 1 | Calculated OHT_1 2 | Assignment |
|---|---|---|
| 1637 | 1626 (374) | NH2 sciss |
| 1595 | 1593 (274) | CC (aniline) stretch + NH2 sciss |
| 1432 | 1457 (118) | NH bend + thiazole breath |
| 1441 (540) | NH bend | |
| 1342 | 1338 (112) | C-NH2 stretch + CH bend |
| 1332 (130) | SO2 stretch asymm + NH bend | |
| 1182 | 1189 (120) | C-S(thiazole) stretch + NH bend |
| 1127 | 1119 (694) | C-SO2 stretch + S-O stretch |
| 1087 | 1069 (67) | S-O stretch + C-SO2 stretch |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Corinti, D.; Chiavarino, B.; Maitre, P.; Crestoni, M.E.; Fornarini, S. Ligation Motifs in Zinc-Bound Sulfonamide Drugs Assayed by IR Ion Spectroscopy. Molecules 2022, 27, 3144. https://doi.org/10.3390/molecules27103144
Corinti D, Chiavarino B, Maitre P, Crestoni ME, Fornarini S. Ligation Motifs in Zinc-Bound Sulfonamide Drugs Assayed by IR Ion Spectroscopy. Molecules. 2022; 27(10):3144. https://doi.org/10.3390/molecules27103144
Chicago/Turabian StyleCorinti, Davide, Barbara Chiavarino, Philippe Maitre, Maria Elisa Crestoni, and Simonetta Fornarini. 2022. "Ligation Motifs in Zinc-Bound Sulfonamide Drugs Assayed by IR Ion Spectroscopy" Molecules 27, no. 10: 3144. https://doi.org/10.3390/molecules27103144
APA StyleCorinti, D., Chiavarino, B., Maitre, P., Crestoni, M. E., & Fornarini, S. (2022). Ligation Motifs in Zinc-Bound Sulfonamide Drugs Assayed by IR Ion Spectroscopy. Molecules, 27(10), 3144. https://doi.org/10.3390/molecules27103144