Peptide Biomarkers Discovery for Seven Species of Deer Antler Using LC-MS/MS and Label-Free Approach
Abstract
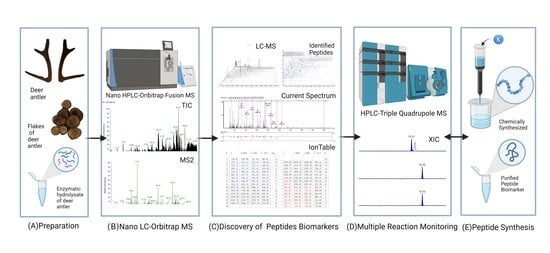
:1. Introduction
2. Results and Discussion
2.1. Results of Nano LC-MS/MS Data Analysis
2.2. Determination and Specificity Verification of Peptide Biomarkers
2.3. Synthesis and Sequence Verification of Peptide Biomarkers
2.4. Strategy 1: Simultaneous Identification of Seven Species of Deer Antlers
2.5. Strategy 2: Identification of Mixtures Such as Deer Antler Flakes or Deer Antler Powder
2.6. Reasons for Choosing HBBA
2.7. Amino Acid Sequence of HBBA in Deer Antler
2.7.1. Amino Acid Sequence of HBBA in Sika Deer Antler
2.7.2. Amino Acid Sequence of HBBA in Sika Deer, Red Deer and North American Wapiti Deer Antler
2.8. Post-Translational Modifications (PTM) of Peptide Biomarkers
3. Materials and Methods
3.1. Materials and Reagents
3.2. Sample Preparation
3.3. Nano LC-MS/MS Analysis
3.4. Mass Spectrometry Data Analysis and Discovery of Peptide Biomarkers
3.5. Verification of the Specificity of Peptide Biomarkers by HPLC-MS/MS
3.6. Synthesis and Verification of Peptide Biomarkers
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Wang, Y.; Zhang, C.; Wang, N.; Li, Z.; Heller, R.; Liu, R.; Zhao, Y.; Han, J.; Pan, X.; Zheng, Z.; et al. Genetic basis of ruminant headgear and rapid antler regeneration. Science 2019, 364, eaav6335. [Google Scholar] [CrossRef] [PubMed]
- Sui, Z.; Zhang, L.; Huo, Y.; Zhang, Y. Bioactive components of velvet antlers and their pharmacological properties. J. Pharm. Biomed. Anal. 2014, 87, 229–240. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.H.; Cheng, X.L.; Liu, W.X.; Li, M.H.; Wei, F.; Ma, S.C. Identification of velvet antler and its mixed varieties by UPLC-QTOF-MS combined with principal component analysis. J. Pharm. Biomed. Anal. 2019, 165, 18–23. [Google Scholar] [CrossRef]
- Sui, Z.; Sun, H.; Weng, Y.; Zhang, X.; Sun, M.; Sun, R.; Zhao, B.; Liang, Z.; Zhang, Y.; Li, C.; et al. Quantitative proteomics analysis of deer antlerogenic periosteal cells reveals potential bioactive factors in velvet antlers. J. Chromatogr. A 2020, 1609, 460496. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Jin, Y.; Wang, F.; Yan, J.; Qi, Y.; Ye, M. Protein digestomic analysis reveals the bioactivity of deer antler velvet in simulated gastrointestinal digestion. Food Res. Int. 2017, 96, 182–190. [Google Scholar] [CrossRef] [PubMed]
- Zheng, K.; Li, Q.; Lin, D.; Zong, X.; Luo, X.; Yang, M.; Yue, X.; Ma, S. Peptidomic analysis of pilose antler and its inhibitory effect on triple-negative breast cancer at multiple sites. Food Funct. 2020, 11, 7481–7494. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Fan, M.; Choi, Y.J.; Yu, Y.; Yao, G.; Deng, Y.; Moon, S.H.; Kim, E.K. Sika deer (Cervus nippon) velvet antler extract attenuates prostate cancer in xenograft model. Biosci. Biotechnol. Biochem. 2019, 83, 348–356. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wang, L.; Wang, J.; Luo, J.; Ruan, H.; Zhang, J. Purified Sika deer antler protein attenuates GM-induced nephrotoxicity by activating Nrf2 pathway and inhibiting NF-kappaB pathway. Sci. Rep. 2020, 10, 15601. [Google Scholar] [CrossRef]
- He, J.; Li, X.; Wang, Z.; Bennett, S.; Chen, K.; Xiao, Z.; Zhan, J.; Chen, S.; Hou, Y.; Chen, J.; et al. Therapeutic anabolic and anticatabolic benefits of natural chinese medicines for the treatment of osteoporosis. Front. Pharmacol. 2019, 10, 1344. [Google Scholar] [CrossRef]
- Zang, Z.J.; Tang, H.F.; Tuo, Y.; Xing, W.J.; Ji, S.Y.; Gao, Y.; Deng, C.H. Effects of velvet antler polypeptide on sexual behavior and testosterone synthesis in aging male mice. Asian J. Androl. 2016, 18, 613–619. [Google Scholar] [CrossRef]
- Xin, J.L.; Zhang, Y.; Li, Y.; Zhang, L.Z.; Lin, Y.; Zheng, L.W. Protective effects of Cervus nippon Temminck velvet antler polypeptides against MPP+induced cytotoxicity in SHSY5Y neuroblastoma cells. Mol. Med. Rep. 2017, 16, 5143–5150. [Google Scholar] [CrossRef] [Green Version]
- Zhao, L.; Mi, Y.; Guan, H.; Xu, Y.; Mei, Y. Velvet antler peptide prevents pressure overload-induced cardiac fibrosis via transforming growth factor (TGF)-beta1 pathway inhibition. Eur. J. Pharmacol. 2016, 783, 33–46. [Google Scholar] [CrossRef]
- Xiao, X.; Xu, S.; Li, L.; Mao, M.; Wang, J.; Li, Y.; Wang, Z.; Ye, F.; Huang, L. The effect of velvet antler proteins on cardiac microvascular endothelial cells challenged with ischemia-hypoxia. Front. Pharmacol. 2017, 8, 601. [Google Scholar] [CrossRef]
- Xiao, X.; Li, L.; Xu, S.; Mao, M.; Pan, R.; Li, Y.; Wu, J.; Huang, L.; Zheng, X. Evaluation of velvet antler total protein effect on bone marrowderived endothelial progenitor cells. Mol. Med. Rep. 2017, 16, 3161–3168. [Google Scholar] [CrossRef] [Green Version]
- Ding, Y.; Ko, S.C.; Moon, S.H.; Lee, S.H. Protective effects of novel antioxidant peptide purified from alcalase hydrolysate of velvet antler against oxidative stress in chang liver cells in vitro and in a zebrafish model in vivo. Int. J. Mol. Sci. 2019, 20, 5187. [Google Scholar] [CrossRef] [Green Version]
- Han, S.; Zhao, K.; Cai, S.; Jiang, M.; Huang, X.; Chen, S.; Li, S.; Zhao, M.; Duan, J.A.; Liu, R. Discovery of peptide biomarkers by label-free peptidomics for discrimination of horn gelatin and hide gelatin from Cervus nippon Temminck. Food Chem. 2021, 363, 130347. [Google Scholar] [CrossRef]
- Batchelder, H.J. Deer Antler Velvet Overview. Available online: https://deerantlervelvet.com/deer-antler-velvet-overview (accessed on 14 June 2022).
- Commission, C.P. Chinese Pharmacopoeia 2020 Edition; Pharmacopoeia of the People’s Republic of China: Beijing, China, 2020; pp. 335–336. [Google Scholar]
- Li, B.; Gao, H.; Song, P.; Liang, C.; Jiang, F.; Xu, B.; Liu, D.; Zhang, T. Captivity shifts gut microbiota communities in white-lipped deer (Cervus albirostris). Animals 2022, 12, 431. [Google Scholar] [CrossRef]
- Wang, L.; Shen, H.; Zheng, Y.; Schumacher, L. Astrovirus in white-tailed deer, United States, 2018. Emerg. Infect. Dis. 2020, 26, 374–376. [Google Scholar] [CrossRef]
- Allen, S.E.; Rothenburger, J.L.; Jardine, C.M.; Ambagala, A.; Hooper-McGrevy, K.; Colucci, N.; Furukawa-Stoffer, T.; Vigil, S.; Ruder, M. Epizootic hemorrhagic disease in white-tailed deer, Canada. Emerg. Infect. Dis. 2019, 25, 832–834. [Google Scholar] [CrossRef] [Green Version]
- Chandler, J.C.; Bevins, S.N.; Ellis, J.W.; Linder, T.J.; Tell, R.M.; Jenkins-Moore, M.; Root, J.J.; Lenoch, J.B.; Robbe-Austerman, S.; DeLiberto, T.J.; et al. SARS-CoV-2 exposure in wild white-tailed deer (Odocoileus virginianus). Proc. Natl. Acad. Sci. USA 2021, 118, e2114828118. [Google Scholar] [CrossRef]
- Szczerba-Turek, A.; Siemionek, J.; Socha, P.; Bancerz-Kisiel, A.; Platt-Samoraj, A.; Lipczynska-Ilczuk, K. Shiga toxin-producing Escherichia coli isolates from red deer (Cervus elaphus), roe deer (Capreolus capreolus) and fallow deer (Dama dama) in Poland. Food Microbiol. 2020, 86, 103352. [Google Scholar] [CrossRef] [PubMed]
- Moraes, D.; Lopez-Lopez, P.; Palmeira, J.D.; Torres, R.T.; Rivero-Juarez, A.; Dutra, V.; Nascimento, M. Screening for hepatitis E virus genotype 3 in red deer (Cervus elaphus) and fallow deer (Dama dama), Portugal, 2018–2020. Transbound. Emerg. Dis. 2021. [Google Scholar] [CrossRef]
- Silaghi, C.; Fröhlich, J.; Reindl, H.; Hamel, D.; Rehbein, S.J. Anaplasma phagocytophilum and Babesia Species of Sympatric Roe Deer (Capreolus capreolus), Fallow Deer (Dama dama), Sika Deer (Cervus nippon) and Red Deer (Cervus elaphus) in Germany. Pathogens 2020, 9, 968. [Google Scholar] [CrossRef]
- Sylvester, K.G.; Ling, X.B.; Liu, G.Y.; Kastenberg, Z.J.; Ji, J.; Hu, Z.; Peng, S.; Lau, K.; Abdullah, F.; Brandt, M.L.; et al. A novel urine peptide biomarker-based algorithm for the prognosis of necrotising enterocolitis in human infants. Gut 2014, 63, 1284–1292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Esin, A.; Bergendahl, L.T.; Savolainen, V.; Marsh, J.A.; Warnecke, T. The genetic basis and evolution of red blood cell sickling in deer. Nat. Ecol. Evol. 2018, 2, 367–376. [Google Scholar] [CrossRef]
- Ba, H.; Wang, D.; Yau, T.O.; Shang, Y.; Li, C. Transcriptomic analysis of different tissue layers in antler growth Center in Sika Deer (Cervus nippon). BMC Genom. 2019, 20, 173. [Google Scholar] [CrossRef] [Green Version]
- Sui, Z.; Weng, Y.; Zhao, Q.; Deng, N.; Fang, F.; Zhu, X.; Shan, Y.; Zhang, L.; Zhang, Y. Ionic liquid-based method for direct proteome characterization of velvet antler cartilage. Talanta 2016, 161, 541–546. [Google Scholar] [CrossRef] [PubMed]
- Hu, P.; Wang, T.; Liu, H.; Xu, J.; Wang, L.; Zhao, P.; Xing, X. Full-length transcriptome and microRNA sequencing reveal the specific gene-regulation network of velvet antler in sika deer with extremely different velvet antler weight. Mol. Genet. Genom. 2019, 294, 431–443. [Google Scholar] [CrossRef]
- Lv, J.J.; Liu, Y.; Zeng, X.Y.; Yu, J.; Li, Y.; Du, X.Q.; Wu, Z.B.; Hao, S.L.; Wang, B.C. Anti-fatigue peptides from the enzymatic hydrolysates of Cervus elaphus blood. Molecules 2021, 26, 7614. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Zidek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold protein structure database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef] [PubMed]










| No. | Sequence | Eurasian Elk | Reindeer | White-Tailed Deer | White-Lipped Deer | Fallow Deer | Sika Deer | Red Deer | Charge | m/z | Production |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Pep 1 | EFTPELQADYQK | √ | 2 | 734.85 | 360.15 | ||||||
| Pep 2 | VDEVGGEALGR | √ | 2 | 551.78 | 659.35 | ||||||
| Pep 3 | FFEHFGDLSTADAVMHNAK | √ | 3 | 713.32 | 880.36 | ||||||
| Pep 4 | VDEVGAEALGR | √ | 2 | 558.29 | 772.43 | ||||||
| Pep 5 | MLTSEEK | √ | 2 | 419.20 | 258.14 | ||||||
| Pep 6 | DFTPVLQADFQK | √ | 2 | 704.86 | 537.27 | ||||||
| Pep 7 | FFEHFGDLSSADAVMGNPK | √ | 3 | 695.64 | 880.36 | ||||||
| Pep 8 | LLGNVLVVVMAR | √ | 2 | 642.39 | 787.49 | ||||||
| Pep 9 | FFEHFGDLSTPDAVMGNPK | √ | √ | 3 | 708.99 | 880.36 | |||||
| Pep 10 | VVAGVANALAHR | √ | √ | √ | 2 | 589.83 | 567.34 | ||||
| Pep 11 | FFEHFGDLSTADAVMGNPK | √ | √ | 3 | 700.32 | 880.36 | |||||
| Pep 12 | VLDAFSDGLK | √ | √ | √ | √ | √ | 2 | 532.78 | 317.22 | ||
| Pep 13 | MLTAEEK | √ | √ | √ | √ | √ | √ | 2 | 411.21 | 258.14 | |
| Pep 14 | AAVTAFWGK | √ | √ | √ | √ | √ | 2 | 475.76 | 390.21 | ||
| Pep 15 | LLGNVLVVVLAR | √ | √ | √ | √ | √ | √ | 2 | 633.42 | 769.53 | |
| Pep 16 | HHGGEFTPVLQADFQK | √ | √ | √ | 3 | 604.30 | 275.13 | ||||
| Pep 17 | AAVTGFWGK | √ | √ | 2 | 468.75 | 567.34 | |||||
| Pep 18 | VLDAFSEGLK | √ | √ | 2 | 539.79 | 866.43 | |||||
| Pep 19 | VVTGVANALAHR | √ | √ | √ | √ | 2 | 604.84 | 567.34 |
| No | Sequence | Modification Site | Types of PTM | Change of Molecular Weight | m/z Before PTM | m/z After PTM |
|---|---|---|---|---|---|---|
| Pep3 | FFEHFGDLSTADAVMHNAK | 17 N | Deamidation | +0.98 | 713.00 | 713.32 |
| Pep7 | FFEHFGDLSSADAVMGNPK | 15 M | Oxidation | +15.99 | 690.65 | 695.64 |
| Pep9 | FFEHFGDLSTPDAVMGNPK | 15 M | Oxidation | +15.99 | 703.99 | 708.99 |
| Pep10 | VVAGVANALAHR | 7 N | Deamidation | +0.98 | 589.34 | 589.83 |
| Pep11 | FFEHFGDLSTADAVMGNPK | 15 M | Oxidation | +15.99 | 694.99 | 700.32 |
| Pep16 | HHGGEFTPVLQADFQK | 14 Q | Deamidation | +0.98 | 604.30 | 604.60 |
| Pep19 | VVTGVANALAHR | 7 N | Deamidation | +0.98 | 604.35 | 604.84 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xue, F.; Wang, B.; Guo, D.-X.; Jiao, Y.; Yin, X.; Cui, W.-L.; Zhou, Q.-Q.; Yu, F.-R.; Lin, Y.-Q. Peptide Biomarkers Discovery for Seven Species of Deer Antler Using LC-MS/MS and Label-Free Approach. Molecules 2022, 27, 4756. https://doi.org/10.3390/molecules27154756
Xue F, Wang B, Guo D-X, Jiao Y, Yin X, Cui W-L, Zhou Q-Q, Yu F-R, Lin Y-Q. Peptide Biomarkers Discovery for Seven Species of Deer Antler Using LC-MS/MS and Label-Free Approach. Molecules. 2022; 27(15):4756. https://doi.org/10.3390/molecules27154756
Chicago/Turabian StyleXue, Fei, Bing Wang, Dong-Xiao Guo, Yang Jiao, Xue Yin, Wei-Liang Cui, Qian-Qian Zhou, Feng-Rui Yu, and Yong-Qiang Lin. 2022. "Peptide Biomarkers Discovery for Seven Species of Deer Antler Using LC-MS/MS and Label-Free Approach" Molecules 27, no. 15: 4756. https://doi.org/10.3390/molecules27154756
APA StyleXue, F., Wang, B., Guo, D. -X., Jiao, Y., Yin, X., Cui, W. -L., Zhou, Q. -Q., Yu, F. -R., & Lin, Y. -Q. (2022). Peptide Biomarkers Discovery for Seven Species of Deer Antler Using LC-MS/MS and Label-Free Approach. Molecules, 27(15), 4756. https://doi.org/10.3390/molecules27154756