Isoquinoline Antimicrobial Agent: Activity against Intracellular Bacteria and Effect on Global Bacterial Proteome
Abstract
:1. Introduction
2. Results and Discussion
2.1. Screening for Antibacterial Active Alkynyl Isoquinolines
2.2. HSN584 and HSN739 Exhibit Broad Spectrum Bactericidal Activity against Gram-Positive Bacteria
2.3. Alkynyl Isoquinolines Are Active against Fluoroquinoline-Resistant Bacteria
2.4. Alkynyl Isoquinolines Intracellular and In Vivo Effects
2.5. S. aureus Does Not Develop Rapid Resistance against HSN584 and HSN789
2.6. Elucidating HSN584 Mechanism of Action
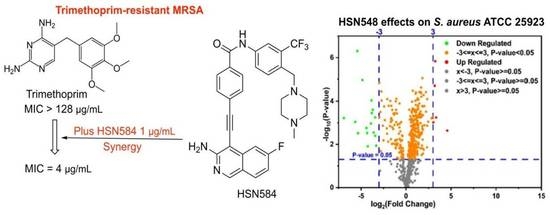
2.7. HSN584 Synergizes with Trimethoprim against Trimethoprim-Resistant MRSA
3. Materials and Methods
3.1. Screening of Isoquinoline Compounds for Antibacterial Activity against S. aureus
3.2. Determination of the MICs and MBCs against Clinically Important Gram-Positive Bacteria
3.3. HSN584 Effects on Membrane Polarization and Membrane Permeation
3.4. In Vitro Cytotoxicity Analysis of HSN Compounds against J774 Cells
3.5. Florescent Imaging
3.6. Examination of Clearance of Intracellular MRSA Present in Murine Macrophage (J774) Cells
3.7. Resistance Selection
3.8. Macromolecule Synthesis Assay
3.9. S. aureus Growth in the Presence of High Salt Concentration
3.10. Global Proteomics
3.11. Checkerboard Assay
3.12. Galleria Mellonella Toxicity Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
Abbreviations
References
- Naclerio, G.A.; Sintim, H.O. Multiple ways to kill bacteria via inhibiting novel cell wall or membrane targets. Future Med. Chem. 2020, 12, 1253–1279. [Google Scholar] [CrossRef] [PubMed]
- Laxminarayan, R.; Van Boeckel, T.; Frost, I.; Kariuki, S.; Khan, E.A.; Limmathurotsakul, D.; Larsson, D.G.J.; Levy-Hara, G.; Mendelson, M.; Outterson, K.; et al. The Lancet Infectious Diseases Commission on antimicrobial resistance: 6 years later. Lancet Infect. Dis. 2020, 20, e51e60. [Google Scholar] [CrossRef]
- Appelbaum, P.C. 2012 and beyond: Potential for the start of a second pre-antibiotic era? J. Antimicrob. Chemother. 2012, 67, 2062–2068. [Google Scholar] [CrossRef] [Green Version]
- Centers for Disease Control and Prevention (CDC). Antibiotic Resistance Threats in the United State; Centers for Disease Control and Prevention (CDC): Fort Collins, CO, USA, 2013.
- Fair, R.J.; Tor, Y. Antibiotics and Bacterial Resistance in the 21st Century. Perspect. Medicinal Chem. 2014, 6, 25–64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ventola, C.L. The Antibiotic Resistance Crisis: Part 1: Causes and Threats. Pharm. Ther. 2015, 40, 277–283. [Google Scholar]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, L.D.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- Fernandes, P. Antibacterial discovery and development—the failure of success? Nat. Biotechnol. 2006, 24, 1497. [Google Scholar] [CrossRef]
- Fernandes, P.; Martens, E. Antibiotics in late clinical development. Biochem. Pharmacol. 2017, 133, 152–163. [Google Scholar] [CrossRef] [Green Version]
- Coates, A.R.M.; Halls, G.; Hu, Y. Novel classes of antibiotics or more of the same? Br. J. Pharmacol. 2011, 163, 184–194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watkins, R.R.; Lemonovich, T.L.; File, T.M. An evidence-based review of linezolid for the treatment of methicillin-resistant Staphylococcus aureus (MRSA): Place in therapy. Core Evid. 2012, 7, 131–143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gu, B.; Kelesidis, T.; Tsiodras, S.; Hindler, J.; Humphries, R.M. The emerging problem of linezolid-resistant Staphylococcus. J. Antimicrob. Chemother. 2013, 68, 4–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thwaites, G.E.; Gant, V. Are bloodstream leukocytes Trojan Horses for the metastasis of Staphylococcus aureus? Nat. Rev. Microbiol. 2011, 9, 215. [Google Scholar] [CrossRef] [PubMed]
- Sandberg, A.; Hessler, J.H.R.; Skov, R.L.; Blom, J.; Frimodt-Møller, N. Intracellular Activity of Antibiotics against Staphylococcus aureus in a Mouse Peritonitis Model. Antimicrob. Agents Chemother. 2009, 53, 1874–1883. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gresham, H.D.; Lowrance, J.H.; Caver, T.E.; Wilson, B.S.; Cheung, A.L.; Lindberg, F.P. Survival of Staphylococcus aureus Inside Neutrophils Contributes to Infection. J. Immunol. 2000, 164, 3713–3722. [Google Scholar] [CrossRef] [Green Version]
- Lehar, S.M.; Pillow, T.; Xu, M.; Staben, L.; Kajihara, K.K.; Vandlen, R.; DePalatis, L.; Raab, H.; Hazenbos, W.L.; Hiroshi Morisaki, J.; et al. Novel antibody–antibiotic conjugate eliminates intracellular S. aureus. Nature 2015, 527, 323–328. [Google Scholar] [CrossRef]
- Barcia-Macay, M.; Seral, C.; Mingeot-Leclercq, M.-P.; Tulkens, P.M.; Van Bambeke, F. Pharmacodynamic evaluation of the intracellular activities of antibiotics against Staphylococcus aureus in a model of THP-1 macrophages. Antimicrob. Agents Chemother. 2006, 50, 841–851. [Google Scholar] [CrossRef] [Green Version]
- Xiong, M.-H.; Li, Y.-J.; Bao, Y.; Yang, X.-Z.; Hu, B.; Wang, J. Bacteria-Responsive Multifunctional Nanogel for Targeted Antibiotic Delivery. Adv. Mater. 2012, 24, 6175–6180. [Google Scholar] [CrossRef]
- O’Connell, K.M.G.; Hodgkinson, J.T.; Sore, H.F.; Welch, M.; Salmond, G.P.C.; Spring, D.R. Combating Multidrug-Resistant Bacteria: Current Strategies for the Discovery of Novel Antibacterials. Angew. Chem. Int. Ed. 2013, 52, 10706–10733. [Google Scholar] [CrossRef] [PubMed]
- Larocque, E.A.; Naganna, N.; Opoku-Temeng, C.; Lambrecht, A.M.; Sintim, H.O. Alkynylnicotinamide-Based Compounds as ABL1 Inhibitors with Potent Activities against Drug-Resistant CML Harboring ABL1(T315I) Mutant Kinase. ChemMedChem 2018, 13, 1172–1180. [Google Scholar] [CrossRef] [PubMed]
- Larocque, E.; Naganna, N.; Ma, X.; Opoku-Temeng, C.; Carter-Cooper, B.; Chopra, G.; Lapidus, R.G.; Sintim, H.O. Aminoisoquinoline benzamides, FLT3 and Src-family kinase inhibitors, potently inhibit proliferation of acute myeloid leukemia cell lines. Future Med. Chem. 2017, 9, 1213–1225. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Zhou, J.; Wang, C.; Carter-Cooper, B.; Yang, F.; Larocque, E.; Fine, J.; Tsuji, G.; Chopra, G.; Lapidus, R.G.; et al. Identification of New FLT3 Inhibitors That Potently Inhibit AML Cell Lines via an Azo Click-It/Staple-It Approach. ACS Med. Chem. Lett. 2017, 8, 492–497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scott, J.D.; Williams, R.M. Chemistry and Biology of the Tetrahydroisoquinoline Antitumor Antibiotics. Chem. Rev. 2002, 102, 1669–1730. [Google Scholar] [CrossRef]
- Galán, A.; Moreno, L.; Párraga, J.; Serrano, Á.; Sanz, M.J.; Cortes, D.; Cabedo, N. Novel isoquinoline derivatives as antimicrobial agents. Bioorg. Med. Chem. 2013, 21, 3221–3230. [Google Scholar] [CrossRef] [PubMed]
- Panchaud, P.; Bruyère, T.; Blumstein, A.-C.; Bur, D.; Chambovey, A.; Ertel, E.A.; Gude, M.; Hubschwerlen, C.; Jacob, L.; Kimmerlin, T.; et al. Discovery and Optimization of Isoquinoline Ethyl Ureas as Antibacterial Agents. J. Med. Chem. 2017, 60, 3755–3775. [Google Scholar] [CrossRef]
- Pérez, A.; Poza, M.; Fernández, A.; del Carmen Fernández, M.; Mallo, S.; Merino, M.; Rumbo-Feal, S.; Cabral, M.P.; Bou, G. Involvement of the AcrAB-TolC Efflux Pump in the Resistance, Fitness, and Virulence of Enterobacter cloacae. Antimicrob. Agents Chemother. 2012, 56, 2084–2090. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tyagi, P.; Singh, M.; Kumari, H.; Kumari, A.; Mukhopadhyay, K. Bactericidal Activity of Curcumin I Is Associated with Damaging of Bacterial Membrane. PLoS ONE 2015, 10, e0121313. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silverman, J.A.; Perlmutter, N.G.; Shapiro, H.M. Correlation of daptomycin bactericidal activity and membrane depolarization in Staphylococcus aureus. Antimicrob. Agents Chemother. 2003, 47, 2538–2544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heeb, S.; Fletcher, M.P.; Chhabra, S.R.; Diggle, S.P.; Williams, P.; Cámara, M. Quinolones: From antibiotics to autoinducers. FEMS Microbiol. Rev. 2011, 35, 247–274. [Google Scholar] [CrossRef] [Green Version]
- Gade, N.D.; Qazi, M.S. Fluoroquinolone Therapy in Staphylococcus aureus Infections: Where Do We Stand? J. Lab. Physicians 2013, 5, 109–112. [Google Scholar] [CrossRef] [PubMed]
- Yamaoka, T. The bactericidal effects of anti-MRSA agents with rifampicin and sulfamethoxazole-trimethoprim against intracellular phagocytized MRSA. J. Infect. Chemother. 2007, 13, 141–146. [Google Scholar] [CrossRef]
- Elsebaei, M.M.; Abutaleb, N.S.; Mahgoub, A.A.; Li, D.; Hagras, M.; Mohammad, H.; Seleem, M.N.; Mayhoub, A.S. Phenylthiazoles with nitrogenous side chain: An approach to overcome molecular obesity. Eur. J. Med. Chem. 2019, 182, 111593. [Google Scholar] [CrossRef] [PubMed]
- Hosny, Y.; Abutaleb, N.S.; Omara, M.; Alhashimi, M.; Elsebaei, M.M.; Elzahabi, H.S.; Seleem, M.N.; Mayhoub, A.S. Modifying the lipophilic part of phenylthiazole antibiotics to control their drug-likeness. Eur. J. Med. Chem. 2020, 185, 111830. [Google Scholar] [CrossRef] [PubMed]
- Elsebaei, M.M.; Mohammad, H.; Samir, A.; Abutaleb, N.S.; Norvil, A.B.; Michie, A.R.; Moustafa, M.M.; Samy, H.; Gowher, H.; Seleem, M.N.; et al. Lipophilic efficient phenylthiazoles with potent undecaprenyl pyrophosphatase inhibitory activity. Eur. J. Med. Chem. 2019, 175, 49–62. [Google Scholar] [CrossRef] [PubMed]
- Joshi, S.; Mumtaz, S.; Singh, J.; Pasha, S.; Mukhopadhyay, K. Novel Miniature Membrane Active Lipopeptidomimetics against Planktonic and Biofilm Embedded Methicillin-Resistant Staphylococcus aureus. Sci. Rep. 2018, 8, 1021. [Google Scholar] [CrossRef] [Green Version]
- Garmory, H.S.; Titball, R.W. ATP-binding cassette transporters are targets for the development of antibacterial vaccines and therapies. Infect. Immun. 2004, 72, 6757–6763. [Google Scholar] [CrossRef] [Green Version]
- Gründling, A. Potassium Uptake Systems in Staphylococcus aureus: New Stories about Ancient Systems. mBio 2013, 4, e00784-13. [Google Scholar] [CrossRef] [Green Version]
- Vermassen, A.; Leroy, S.; Talon, R.; Provot, C.; Popowska, M.; Desvaux, M. Cell Wall Hydrolases in Bacteria: Insight on the Diversity of Cell Wall Amidases, Glycosidases and Peptidases Toward Peptidoglycan. Front. Microbiol. 2019, 10, 331. [Google Scholar] [CrossRef]
- Connelly, J.C.; Kirkham, L.A.; Leach, D.R.F. The SbcCD nuclease of Escherichia coli is a structural maintenance of chromosomes (SMC) family protein that cleaves hairpin DNA. Proc. Nat. Acad. Sci. USA 1998, 95, 7969–7974. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fowler, R.G.; Schaaper, R.M. The role of the mutT gene of Escherichia coli in maintaining replication fidelity. FEMS Microbiol. Rev. 1997, 21, 43–54. [Google Scholar] [CrossRef] [Green Version]
- Ho, H.N.; van Oijen, A.M.; Ghodke, H. The transcription-repair coupling factor Mfd associates with RNA polymerase in the absence of exogenous damage. Nat. Commun. 2018, 9, 1570. [Google Scholar] [CrossRef] [Green Version]
- Deaconescu, A.M.; Savery, N.; Darst, S.A. The bacterial transcription repair coupling factor. Curr. Opin. Struct. Biol. 2007, 17, 96–102. [Google Scholar] [CrossRef] [Green Version]
- Gregersen, L.H.; Svejstrup, J.Q. The Cellular Response to Transcription-Blocking DNA Damage. Trends Biochem. Sci. 2018, 43, 327–341. [Google Scholar] [CrossRef]
- Elmwall, J.; Kwiecinski, J.; Na, M.; Ali, A.A.; Osla, V.; Shaw, L.N.; Wang, W.; Sävman, K.; Josefsson, E.; Bylund, J.; et al. Galectin-3 Is a Target for Proteases Involved in the Virulence of Staphylococcus aureus. Infect. Immun. 2017, 85, e00177-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, L.Y.; Miyamoto, Y.J.; McIntyre, B.W.; Höök, M.; McCrea, K.W.; McDevitt, D.; Brown, E.L. The Staphylococcus aureus Map protein is an immunomodulator that interferes with T cell-mediated responses. J. Clin. Investig. 2002, 110, 1461–1471. [Google Scholar] [CrossRef] [PubMed]
- Smagur, J.; Guzik, K.; Magiera, L.; Bzowska, M.; Gruca, M.; Thøgersen, I.B.; Enghild, J.J.; Potempa, J. A new pathway of staphylococcal pathogenesis: Apoptosis-like death induced by Staphopain B in human neutrophils and monocytes. J. Innate Immun. 2009, 1, 98–108. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, I.M.; Hartford, O.; Foster, T.; Tarkowski, A. Alpha-toxin and gamma-toxin jointly promote Staphylococcus aureus virulence in murine septic arthritis. Infect. Immun. 1999, 67, 1045–1049. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bouza, E.; Muñoz, P. Monotherapy versus combination therapy for bacterial infections. Med. Clin. N. Am. 2000, 84, 1357–1389. [Google Scholar] [CrossRef]
- Orhan, G.; Bayram, A.; Zer, Y.; Balci, I. Synergy Tests by E Test and Checkerboard Methods of Antimicrobial Combinations against Brucella melitensis. J. Clin. Microbiol. 2005, 43, 140–143. [Google Scholar] [CrossRef] [Green Version]
- CLSI Document M07-A9; Methods for Dilution Antimicrobial Susceptibility Tests f or Bacteria That Grow Aerobically. Approved Standard—Ninth Edition; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2012.
- Abutaleb, N.S.; Seleem, M.N. Repurposing the Antiamoebic Drug Diiodohydroxyquinoline for Treatment of Clostridioides difficile Infections. Antimicrob. Agents Chemother. 2020, 64, e02115–e02119. [Google Scholar] [CrossRef] [PubMed]
- Naclerio, G.A.; Abutaleb, N.S.; Li, D.; Seleem, M.N.; Sintim, H.O. Ultrapotent Inhibitor of Clostridioides difficile Growth, Which Suppresses Recurrence In Vivo. J.Med. Chem. 2020, 63, 11934–11944. [Google Scholar] [CrossRef]
- Abutaleb, N.S.; Seleem, M.N. Auranofin, at clinically achievable dose, protects mice and prevents recurrence from Clostridioides difficile infection. Sci. Rep. 2020, 10, 7701. [Google Scholar] [CrossRef] [PubMed]
- Abutaleb, N.S.; Seleem, M.N. Antivirulence activity of auranofin against vancomycin-resistant enterococci: In vitro and in vivo studies. Int. J. Antimicrob. Agents 2020, 55, 105828. [Google Scholar] [CrossRef] [PubMed]
- Kotb, A.; Abutaleb, N.S.; Hagras, M.; Bayoumi, A.; Moustafa, M.M.; Ghiaty, A.; Seleem, M.N.; Mayhoub, A.S. tert-Butylphenylthiazoles with an oxadiazole linker: A novel orally bioavailable class of antibiotics exhibiting antibiofilm activity. RSC Advances 2019, 9, 6770–6778. [Google Scholar] [CrossRef] [Green Version]
- Mancy, A.; Abutaleb, N.S.; Elsebaei, M.M.; Saad, A.Y.; Kotb, A.; Ali, A.O.; Abdel-Aleem, J.A.; Mohammad, H.; Seleem, M.N.; Mayhoub, A.S. Balancing Physicochemical Properties of Phenylthiazole Compounds with Antibacterial Potency by Modifying the Lipophilic Side Chain. ACS Infect. Dis. 2020, 6, 80–90. [Google Scholar] [CrossRef] [PubMed]
- Shahin, I.G.; Abutaleb, N.S.; Alhashimi, M.; Kassab, A.E.; Mohamed, K.O.; Taher, A.T.; Seleem, M.N.; Mayhoub, A.S. Evaluation of N-phenyl-2-aminothiazoles for treatment of multi-drug resistant and intracellular Staphylococcus aureus infections. Eur. J. Med. Chem. 2020, 202, 112497. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Hancock, R.E.W. Interaction of the Cyclic Antimicrobial Cationic Peptide Bactenecin with the Outer and Cytoplasmic Membrane. J. Biol. Chem. 1999, 274, 29–35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yasir, M.; Dutta, D.; Willcox, M.D.P. Mode of action of the antimicrobial peptide Mel4 is independent of Staphylococcus aureus cell membrane permeability. PLoS ONE 2019, 14, e0215703. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Shi, Y.; Su, G.; Le, G. Selectivity for and destruction of Salmonella typhimurium via a membrane damage mechanism of a cell-penetrating peptide ppTG20 analogue. Int. J. Antimicrob. Agents 2012, 40, 337–343. [Google Scholar] [CrossRef] [PubMed]
- Naclerio, G.A.; Abutaleb, N.S.; Onyedibe, K.I.; Seleem, M.N.; Sintim, H.O. Potent trifluoromethoxy, trifluoromethylsulfonyl, trifluoromethylthio and pentafluorosulfanyl containing (1,3,4-oxadiazol-2-yl)benzamides against drug-resistant Gram-positive bacteria. RSC Med. Chem. 2019, 11, 102–110. [Google Scholar] [CrossRef]
- Hagras, M.; Abutaleb, N.S.; Ali, A.O.; Abdel-Aleem, J.A.; Elsebaei, M.M.; Seleem, M.N.; Mayhoub, A.S. Naphthylthiazoles: Targeting Multidrug-Resistant and Intracellular Staphylococcus aureus with Biofilm Disruption Activity. ACS Infect. Dis. 2018, 4, 1679–1691. [Google Scholar] [CrossRef]
- Dayal, N.; Opoku-Temeng, C.; Mohammad, H.; Abutaleb, N.S.; Hernandez, D.; Onyedibe, K.I.; Wang, M.; Zeller, M.; Seleem, M.N.; Sintim, H.O. Inhibitors of Intracellular Gram-Positive Bacterial Growth Synthesized via Povarov-Doebner Reactions. ACS Infect. Dis. 2019, 5, 1820–1830. [Google Scholar] [CrossRef]
- Singh, M.P.; Petersen, P.J.; Weiss, W.J.; Janso, J.E.; Luckman, S.W.; Lenoy, E.B.; Bradford, P.A.; Testa, R.T.; Greenstein, M. Mannopeptimycins, new cyclic glycopeptide antibiotics produced by Streptomyces hygroscopicus LL-AC98: Antibacterial and mechanistic activities. Antimicrob. Agents Chemother. 2003, 47, 62–69. [Google Scholar] [CrossRef] [Green Version]
- Opoku-Temeng, C.; Naclerio, G.A.; Mohammad, H.; Dayal, N.; Abutaleb, N.S.; Seleem, M.N.; Sintim, H.O. N-(1,3,4-oxadiazol-2-yl)benzamide analogs, bacteriostatic agents against methicillin- and vancomycin-resistant bacteria. Eur. J. Med. Chem. 2018, 155, 797–805. [Google Scholar] [CrossRef] [PubMed]
- Opoku-Temeng, C.; Onyedibe, K.I.; Aryal, U.K.; Sintim, H.O. Proteomic analysis of bacterial response to a 4-hydroxybenzylidene indolinone compound, which re-sensitizes bacteria to traditional antibiotics. J. Proteom. 2019, 202, 103368. [Google Scholar] [CrossRef] [PubMed]
- Eldesouky, H.E.; Li, X.; Abutaleb, N.S.; Mohammad, H.; Seleem, M.N. Synergistic interactions of sulfamethoxazole and azole antifungal drugs against emerging multidrug-resistant Candida auris. Int. J. Antimicrob. Agents 2018, 52, 754–761. [Google Scholar] [CrossRef]
- Mohammad, H.; Cushman, M.; Seleem, M.N. Antibacterial Evaluation of Synthetic Thiazole Compounds In Vitro and In Vivo in a Methicillin-Resistant Staphylococcus aureus (MRSA) Skin Infection Mouse Model. PLoS ONE 2015, 10, e0142321. [Google Scholar] [CrossRef] [Green Version]
- Meletiadis, J.; Pournaras, S.; Roilides, E.; Walsh, T.J. Defining Fractional Inhibitory Concentration Index Cutoffs for Additive Interactions Based on Self-Drug Additive Combinations, Monte Carlo Simulation Analysis, and In Vitro-In Vivo Correlation Data for Antifungal Drug Combinations against Aspergillus fumigatus. Antimicrob. Agents Chemother. 2010, 54, 602. [Google Scholar] [PubMed] [Green Version]
- Megaw, J.; Thompson, T.P.; Lafferty, R.A.; Gilmore, B.F. Galleria mellonella as a novel in vivo model for assessment of the toxicity of 1-alkyl-3-methylimidazolium chloride ionic liquids. Chemosphere 2015, 139, 197–201. [Google Scholar] [CrossRef] [PubMed]
- Harding, C.R.; Schroeder, G.N.; Collins, J.W.; Frankel, G. Use of Galleria mellonella as a model organism to study Legionella pneumophila infection. J. Vis. Exp. 2013, 81, e50964. [Google Scholar]










| S. aureus ATCC 25923 (µg/mL) | MRSA ATCC 33592 (µg/mL) | MRSA USA 300-0114 (µg/mL) | E. faecalis (µg/mL) | VRE (µg/mL) | |
|---|---|---|---|---|---|
| HSN584 | 2 | 2 | 2 | 2 | 2 |
| HSN738 | 8 | 8 | 4 | 2 | 8 |
| HSN739 | 2 | 2 | 2 | 1 | 2 |
| HSN585 | 4 | 4 | 4 | 4 | 4 |
| Bacterial Strains | Compounds/Control Drugs | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HSN 490 | HSN 584 | HSN 739 | Linezolid | Ciprofloxacin | Levofloxacin | Norfloxacin | ||||||||
| MIC | MBC | MIC | MBC | MIC | MBC | MIC | MBC | MIC | MBC | MIC | MBC | MIC | MBC | |
| MRSA NRS 385 (USA 500) | 8 | 16 | 4 | 4 | 8 | 8 | 2 | 32 | >128 | >128 | 32 | 32 | >128 | >128 |
| MRSA NRS 383 (USA 200) | 16 | 32 | 8 | 8 | 8 | 8 | 2 | 32 | 128 | 128 | 32 | 32 | >128 | >128 |
| MRSA NRS 382 (USA 100) | 16 | 32 | 4 | 4 | 8 | 8 | 2 | 64 | 32 | 32 | 16 | 16 | 128 | 128 |
| VRSA 4 | 16 | 16 | 4 | 4 | 8 | 8 | 1 | 16 | 128 | 128 | 32 | 32 | >128 | >128 |
| VRSA 7 | 16 | 16 | 4 | 4 | 4 | 4 | 1 | 16 | 128 | 128 | 32 | 32 | >128 | >128 |
| VRSA 8 | 16 | 32 | 4 | 4 | 4 | 4 | 0.5 | 16 | 128 | 128 | 32 | 32 | 128 | 128 |
| VRSA 9 | 16 | 16 | 4 | 8 | 4 | 8 | 1 | 64 | >128 | >128 | 32 | 32 | >128 | >128 |
| VRSA 10 | 16 | 16 | 4 | 8 | 8 | 8 | 1 | 64 | >128 | >128 | 32 | 64 | >128 | >128 |
| VRSA 13 | 16 | 32 | 4 | 4 | 4 | 4 | 1 | 64 | 128 | 128 | 16 | 16 | >128 | >128 |
| Protein Name | Average MS/MS Count |
|---|---|
| DUF5067 domain-containing protein | 73 |
| Multidrug transporter | 38 |
| Nickel ABC transporter | 35 |
| Peptide ABC transporter substrate-binding protein | 30 |
| Phosphonate ABC transporter substrate-binding protein | 23 |
| Osmoprotectant ABC transporter substrate-binding protein | 21 |
| Tandem-type lipoprotein | 19 |
| Staphopain B | 17 |
| Protein EssB | 14 |
| Gamma-hemolysin component C | 13 |
| Glycerophosphodiester Phosphodiesterase | 11 |
| Peptidase M23B | 10 |
| Iron ABC transporter substrate-binding protein | 10 |
| Teichoic acid ABC transporter ATP-binding protein | 7 |
| Polysaccharide deacetylase | 7 |
| Sodium ABC transporter permease | 6 |
| Cell division protein FtsQ | 6 |
| Peptide ABC transporter permease | 5 |
| Nitrate ABC transporter substrate-binding protein | 5 |
| Protein Name | Average MS/MS Count |
|---|---|
| Ribosome silencing factor RsfS | 28 |
| DNA mismatch repair protein MutT | 13 |
| XRE family transcriptional regulator | 13 |
| Mevalonate kinase | 12 |
| Ribosome maturation factor | 12 |
| DNA repair protein RadA | 12 |
| Shikimate dehydrogenase | 12 |
| Betaine-aldehyde dehydrogenase | 12 |
| Dihydroxyacetone kinase subunit L | 11 |
| Glycine cleavage system protein H | 11 |
| Endoribonuclease YbeY | 11 |
| Molybdoprotein synthase sulfur carrier subunit | 11 |
| Phosphomevalonate kinase | 10 |
| Cell division protein FtsK | 10 |
| Transcriptional-repair coupling factor | 10 |
| tRNA pseudouridine(55) synthase TruB | 10 |
| Ketol-acid reductoisomerase | 10 |
| Two-component sensor histidine kinase | 10 |
| HAD family hydrolase | 10 |
| FAD-dependent oxidoreductase | 9 |
| Signal peptidase II | 9 |
| Type II secretion protein | 9 |
| 3-dehydroquinate synthase | 9 |
| DNA polymerase/3-5 exonuclease PolX | 9 |
| Polyisoprenoid-binding protein | 9 |
| Cytidine deaminase | 9 |
| Acylphosphatase | 9 |
| Initiation-control protein YabA | 9 |
| Exodeoxyribonuclease 7 large subunit | 8 |
| Isochorismate synthase | 8 |
| LytTR family transcriptional regulator | 8 |
| Cell division protein ZapA | 8 |
| Nuclease SbcCD subunit C | 7 |
| Holliday junction resolvase RuvX | 7 |
| 3-phosphoshikimate 1-carboxyvinyltransferase | 7 |
| Holliday junction branch migration protein RuvA | 7 |
| DNA methyltransferase | 7 |
| DNA mismatch repair protein MutL | 6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karanja, C.W.; Naganna, N.; Abutaleb, N.S.; Dayal, N.; Onyedibe, K.I.; Aryal, U.; Seleem, M.N.; Sintim, H.O. Isoquinoline Antimicrobial Agent: Activity against Intracellular Bacteria and Effect on Global Bacterial Proteome. Molecules 2022, 27, 5085. https://doi.org/10.3390/molecules27165085
Karanja CW, Naganna N, Abutaleb NS, Dayal N, Onyedibe KI, Aryal U, Seleem MN, Sintim HO. Isoquinoline Antimicrobial Agent: Activity against Intracellular Bacteria and Effect on Global Bacterial Proteome. Molecules. 2022; 27(16):5085. https://doi.org/10.3390/molecules27165085
Chicago/Turabian StyleKaranja, Caroline W., Nimishetti Naganna, Nader S. Abutaleb, Neetu Dayal, Kenneth I. Onyedibe, Uma Aryal, Mohamed N. Seleem, and Herman O. Sintim. 2022. "Isoquinoline Antimicrobial Agent: Activity against Intracellular Bacteria and Effect on Global Bacterial Proteome" Molecules 27, no. 16: 5085. https://doi.org/10.3390/molecules27165085
APA StyleKaranja, C. W., Naganna, N., Abutaleb, N. S., Dayal, N., Onyedibe, K. I., Aryal, U., Seleem, M. N., & Sintim, H. O. (2022). Isoquinoline Antimicrobial Agent: Activity against Intracellular Bacteria and Effect on Global Bacterial Proteome. Molecules, 27(16), 5085. https://doi.org/10.3390/molecules27165085