Two Novel Sesquiterpenoid Glycosides from the Rhizomes of Atractylodes lancea
Abstract
:1. Introduction
2. Results and Discussion
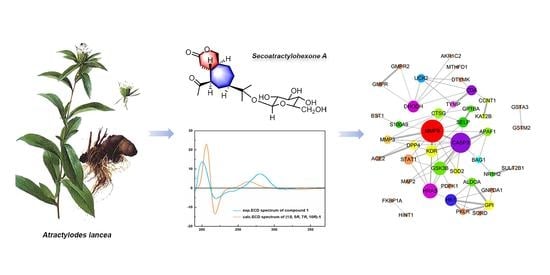
2.1. Structural Elucidation of the Isolated Compounds
2.2. Network Pharmacology-Based Prediction of the Potential Biological Activity of Secoatractylohexone A
3. Materials and Methods
3.1. General Procedures
3.2. Plant Material
3.3. Extraction and Isolation
3.4. General Method for Acid Hydrolysis
3.5. Quantum Chemical Calculation
3.6. Network Pharmacology-Based Prediction of the Potential Biological Actions of Compound 1
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Commission, C.P. Pharmacopoeia of China; Chemical Industry Press: Beijing, China, 2020; pp. 168–169. [Google Scholar]
- Wang, H.X.; Liu, C.M.; Liu, Q.; Gao, K. Three types of sesquiterpenes from rhizomes of Atractylodes lancea. Phytochemistry 2008, 69, 2088–2094. [Google Scholar] [CrossRef]
- Yin, M.; Xiao, C.C.; Chen, Y.; Ming, W.; Guan, F.Q.; Dong, Y.F.; Feng, X. Two New Sesquiterpenoid Glycosides from Rhizomes of Atractylodes lancea. Chem. Nat. Compd. 2015, 51, 495–499. [Google Scholar] [CrossRef]
- Yin, M.; Xiao, C.C.; Chen, Y.; Wang, M.; Guan, F.Q.; Wang, Q.Z.; Shan, Y.; Feng, X. A New Sesquiterpenoid Glycoside from Rhizomes of Atractylodes lancea. Chin. Herb. Med. 2015, 7, 371–374. [Google Scholar] [CrossRef]
- Xu, K.; Feng, Z.M.; Yang, Y.N.; Jiang, J.S.; Zhang, P.C. Eight new eudesmane- and eremophilane-type sesquiterpenoids from Atractylodes lancea. Fitoterapia 2016, 114, 115–121. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.; Jiang, J.S.; Feng, Z.M.; Yang, Y.N.; Li, L.; Zang, C.X.; Zhang, P.C. Bioactive Sesquiterpenoid and Polyacetylene Glycosides from Atractylodes lancea. J. Nat. Prod. 2016, 79, 1567–1575. [Google Scholar] [CrossRef]
- Xu, K.; Feng, Z.M.; Jiang, J.S.; Yang, Y.N.; Zhang, P.C. Sesquiterpenoid and C 14 -polyacetylene glycosides from the rhizomes of Atractylodes lancea. Chin. Chem. Lett. 2017, 28, 597–601. [Google Scholar] [CrossRef]
- Long, L.; Wang, L.; Qi, S.; Yang, Y.; Gao, H. New sesquiterpenoid glycoside from the rhizomes of Atractylodes lancea. Nat. Prod. Res. 2020, 34, 1138–1145. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Zhang, Y.; Peng, X.; Huang, S.; Zhou, H.; Xu, J.; Gu, Q. Diverse Sesquiterpenoids and polyacetylenes from Atractylodes lancea and their anti-osteoclastogenesis activity. J. Nat. Prod. 2022, 85, 866–877. [Google Scholar] [CrossRef]
- Ji, Y.; Feng, X.; Xiao, C.C.; Dong, Y.F.; Wang, Q.Z.; Wang, M.; Zhao, Y.Y. A new polyacetylene glycoside from the rhizomes of Atractylodes lancea. Chin. Chem. Lett. 2010, 21, 850–852. [Google Scholar] [CrossRef]
- Feng, Z.M.; Xu, K.; Wang, W.; Du, N.; Zhang, J.H.; Yang, Y.N.; Jiang, J.S.; Zhang, P.C. Two new thiophene polyacetylene glycosides from Atractylodes lancea. J. Asian Nat. Prod. Res. 2018, 20, 531–537. [Google Scholar] [CrossRef]
- Xu, K.; Feng, Z.M.; Yang, Y.N.; Jiang, J.S.; Zhang, P.C. Four new C-10-polyacetylene glycosides from the rhizomes of Atractylodes lancea. J. Asian Nat. Prod. Res. 2017, 19, 121–127. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.Y.; Zhuang, D.; Wang, H.Y.; Liu, C.Y.; Lv, G.P.; Meng, L.J. Preparation, characterization, and bioactivity evaluation of oligosaccharides from Atractylodes lancea (Thunb.) DC. Carbohydr Polym. 2022, 277, 118854. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.J.; Zhao, Z.Y.; Chang, L.K.; Cao, Y.; Wang, S.; Kang, C.Z.; Wang, H.Y.; Zhou, L.; Huang, L.Q.; Guo, L.P. Atractylodis Rhizoma: A review of its traditional uses, phytochemistry, pharmacology, toxicology and quality control. J. Ethnopharmacol. 2021, 266, 113415. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Wang, C.; Guo, Y.; Wen, R.; Yan, L.; Zhang, F.; Gong, Q.; Yu, H. Research on the effect and underlying molecular mechanism of Cangzhu in the treatment of gouty arthritis. Eur. J. Pharmacol. 2022, 927, 175044. [Google Scholar] [CrossRef]
- Han, J.; Li, W.; Shi, G.; Huang, Y.; Sun, X.; Sun, N.; Jiang, D. Atractylenolide III improves mitochondrial function and protects against ulcerative colitis by activating AMPK/SIRT1/PGC-1alpha. Mediators Inflamm. 2022, 2022, 9129984. [Google Scholar] [CrossRef]
- Wang, J.; Nasser, M.I.; Adlat, S.; Ming Jiang, M.; Jiang, N.; Gao, L. Atractylenolide II induces apoptosis of prostate cancer cells through regulation of AR and JAK2/STAT3 signaling pathways. Molecules 2018, 23, 3298. [Google Scholar] [CrossRef]
- Li, H.; Wang, F.; Zhou, Z.; Jiang, X.; Li, F.; Feng, Y.; Liu, C.; Zhang, Y.; Fan, S.; Wu, X.; et al. Atractylon, a novel dopamine 2 receptor agonist, ameliorates Parkinsonian like motor dysfunctions in MPTP-induced mice. NeuroToxicology 2022, 89, 121–126. [Google Scholar] [CrossRef]
- Du, Z.; Ma, Z.; Lai, S.; Ding, Q.; Hu, Z.; Yang, W.; Qian, Q.; Zhu, L.; Dou, X.; Li, S. Atractylenolide I ameliorates acetaminophen-induced acute liver injury via the TLR4/MAPKs/NF-kappaB signaling pathways. Front. Pharmacol. 2022, 13, 797499. [Google Scholar] [CrossRef]
- Li, H.; Cao, W.; Zhang, X.B.; Zhang, X.X.; Gu, C.; Gu, L.M.; Pan, C.Y.; Tian, Y.Z.; Lu, M. Atractylenolide1 alleviates gastroparesis in diabetic rats by activating the stem cell factor/ckit signaling pathway. Mol. Med. Rep. 2021, 24, 691. [Google Scholar] [CrossRef]
- Li, S.; Zhang, B. Traditional Chinese medicine network pharmacology: Theory, methodology and application. Chin. J. Nat. Med. 2013, 11, 110–120. [Google Scholar] [CrossRef]
- Xu, T.; Li, S.; Sun, Y.; Pi, Z.; Liu, S.; Song, F.; Liu, Z. Systematically characterize the absorbed effective substances of Wutou Decoction and their metabolic pathways in rat plasma using UHPLC-Q-TOF-MS combined with a target network pharmacological analysis. J. Pharm. Biomed. Anal. 2017, 141, 95–107. [Google Scholar] [CrossRef]
- Tang, H.; He, S.; Zhang, X.; Luo, S.; Zhang, B.; Duan, X.; Zhang, Z.; Wang, W.; Wang, Y.; Sun, Y. A network pharmacology approach to uncover the pharmacological mechanism of XuanHuSuo powder on osteoarthritis. Evid. Based Complement. Alternat. Med. 2016, 2016, 3246946. [Google Scholar] [CrossRef]
- Yahara, S.; Higashi, T.; Iwaki, K.; Nohara, T.; Marubayashi, N.; Ueda, I.; Kohda, H.; GOTO, K.; Izumi, H.; Nuno, M.; et al. Study on the constituents of Atractylodes lancea. Chem. Pharm. Bull. 1989, 37, 2995–3000. [Google Scholar] [CrossRef]
- Kitajima, J.; Kamoshita, A.; Ishikawa, T.; Takano, A.; Fukuda, T.; Isoda, S.; Ida, Y. Glycosides of Atractylodes japoniea. Chem. Pharm. Bull. 2003, 51, 152–157. [Google Scholar] [CrossRef] [PubMed]
- Kibble, M.; Saarinen, N.; Tang, J.; Wennerberg, K.; Makela, S.; Aittokallio, T. Network pharmacology applications to map the unexplored target space and therapeutic potential of natural products. Nat. Prod. Rep. 2015, 32, 1249–1266. [Google Scholar] [CrossRef] [PubMed]
- Spartan 14; Wavefunction: Irvine, CA, USA, 2014.
- GAUSSIAN 09, Revision A.1; Gaussian: Wallingford, CT, USA, 2009.
- Bruhn, T.; Schaumlöffel, A.; Hemberger, Y.; Bringmann, G. SpecDis: Quantifying the comparison of calculated and experimental electronic circular dichroism spectra. Chirality 2013, 25, 243–249. [Google Scholar] [CrossRef]



| No. | δC | δH (J, Hz) | HMBC | ROESY |
|---|---|---|---|---|
| 1 | 40.3 | 2.21, m, 1H | C-2, 4, 5, 9, 10, 14 | H-14, 15 |
| 2 | 36.0 | 2.48, dd (1H, 10.6, 17.8), H-2ax | C-1, 3, 5, 10 | H-6, 9, 10, 15 |
| 2.86, dd (1H, 6.1, 17.8), H-2eq | ||||
| 3 | 170.3 | |||
| 4 | 211.2 | |||
| 5 | 59.8 | 3.06, m, 1H | C-1, 2, 4, 6, 7, 10 | H-10, 15 |
| 6 | 28.3 | 1.21, m, 1H, H-6ax | C-1, 5, 7, 8 | H-2, 15 |
| 2.43, m, 1H, H-6eq | C-4, 11 | |||
| 7 | 46.2 | 2.22, m, 1H | C-5, 11 | H-1′, 10, 12, 13 |
| 8 | 26.9 | 1.22, m, 1H, H-8ax | C-9 | H-12, 13 |
| 1.59, m, 1H, H-8eq | ||||
| 9 | 27.0 | 1.20, m, 2H | C-1, 8, 14 | H-2, 14 |
| 10 | 35.6 | 1.69, m, 1H | C-8, 14 | H-2eq, 5, 7 |
| 11 | 80.3 | |||
| 12 | 21.7 | 1.16, s, CH3 | C-3′, 5′, 11, 7, 13 | H-1′, 7, 8 |
| 13 | 25.1 | 1.42, s, CH3 | C-2′, 3′, 4′, 5′, 7, 11, 12 | H-1′, 7, 8 |
| 14 | 73.7 | 3.76, dd (1H, 11.2, 11.3), H-14ax | C-1, 3, 10 | H-1, 9 |
| 4.05, dd (1H, 9.0, 11.2), H-14eq | ||||
| 15 | 29.7 | 2.30, s, CH3 | C-4, 5 | H-1, 2, 5, 6 |
| 1′ | 98.8 | 5.03, d (1H, 7.7) | C-11 | H-7, 12, 13 |
| 2′ | 75.5 | 3.95, m, 1H | C-1′, 3′ | |
| 3′ | 78.9 | 4.20, m, 1H | C-2′, 4′ | |
| 4′ | 72.1 | 4.06, m, 1H | C-3′, 5′, 6′ | |
| 5′ | 78.8 | 3.97, m, 1H | C-4′ | |
| 6′ | 63.0 | 4.62, dd (1H, 1.9, 11.4), H-6′ax | C-4′, 5′ | |
| 4.24, br d (1H, 11.4), H-6′eq |
| No. | δC | δH (J, Hz) | HMBC | ROESY |
|---|---|---|---|---|
| 1 | 42.1 | 2.91, m, 1H | C-2, 5, 9, 10 | H-1, 9, 14 |
| 2 | 40.7 | 2.75, dd (1H, 9.5, 11.8), H-2ax | C-1, 3, 5, 10 | H-4, 5, 8, 14 |
| 3.03, dd (1H, 6.1,11.8), H-2eq | ||||
| 3 | 219.1 | |||
| 4 | 52.1 | 1.79, m, 1H | C-3, 5, 6, 15 | H-2, 15 |
| 5 | 49.4 | 1.57, m, 1H | H-1, 2, 7, 15 | |
| 6 | 39.4 | 1.26, br dd (1H, 12.6, 11.8), H-6ax | C-1, 5, 7, 8 | H-8, 12, 13, 15 |
| 2.61, br d (1H, 12.6), H-6eq | ||||
| 7 | 46.2 | 1.76, m, 1H | C-6, 8, 11, 12, 13 | H-1′, 5, 12, 13 |
| 8 | 29.4 | 2.05, m, 1H, H-8ax | C-6, 7, 9, 10, 11 | H-2, 6, 12, 13 |
| 2.91, m, 1H, H-8eq | ||||
| 9 | 129.9 | 6.24, d, (1H, 7.0) | C-1, 8, 14 | H-1, 14 |
| 10 | 143.9 | |||
| 11 | 80.0 | |||
| 12 | 23.9 | 1.39, s, CH3 | C-7, 11, 13 | H-1′, 6, 7, 8 |
| 13 | 24.0 | 1.40, s, CH3 | C-7, 11, 12 | H-1′, 6, 7, 8 |
| 14 | 66.1 | 4.28, br s, 2H | C-1, 9, 10 | H-1, 2, 9 |
| 15 | 12.8 | 1.10, d (3H, 6.9), CH3 | C-3, 4, 5 | H-4, 5, 6 |
| 1′ | 98.6 | 4.99, d (1H, 7.7) | C-11, 3′ | H-7, 12, 13 |
| 2′ | 75.3 | 3.97, dd (1H, 8.0, 7.8) | C-1′, 3′ | 2′-OH |
| 3′ | 78.9 | 4.21, dd ( 1H, 8.6, 7.7) | C-2′, 4′ | |
| 4′ | 71.9 | 4.18, dd (1H, 7.7, 8.7) | C-3′, 5′ | |
| 5′ | 78.0 | 3.87, m, 1H | C-4′ | |
| 6′ | 63.0 | 4.31, dd (1H, 5.3, 11.5), H-6′ax | C-4′, 5′ | |
| 4.46, br d (1H, 11.5), H-6′eq |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, L.; Guan, F.; Chen, Y.; Wang, F.; Chen, P.; Yin, M.; Wang, B.; Li, L.; Wang, Q.; Gu, Y.; et al. Two Novel Sesquiterpenoid Glycosides from the Rhizomes of Atractylodes lancea. Molecules 2022, 27, 5753. https://doi.org/10.3390/molecules27185753
Liu L, Guan F, Chen Y, Wang F, Chen P, Yin M, Wang B, Li L, Wang Q, Gu Y, et al. Two Novel Sesquiterpenoid Glycosides from the Rhizomes of Atractylodes lancea. Molecules. 2022; 27(18):5753. https://doi.org/10.3390/molecules27185753
Chicago/Turabian StyleLiu, Lanying, Fuqin Guan, Yu Chen, Fan Wang, Pengxu Chen, Min Yin, Bi Wang, Linwei Li, Qizhi Wang, Yonghua Gu, and et al. 2022. "Two Novel Sesquiterpenoid Glycosides from the Rhizomes of Atractylodes lancea" Molecules 27, no. 18: 5753. https://doi.org/10.3390/molecules27185753
APA StyleLiu, L., Guan, F., Chen, Y., Wang, F., Chen, P., Yin, M., Wang, B., Li, L., Wang, Q., Gu, Y., & Feng, X. (2022). Two Novel Sesquiterpenoid Glycosides from the Rhizomes of Atractylodes lancea. Molecules, 27(18), 5753. https://doi.org/10.3390/molecules27185753