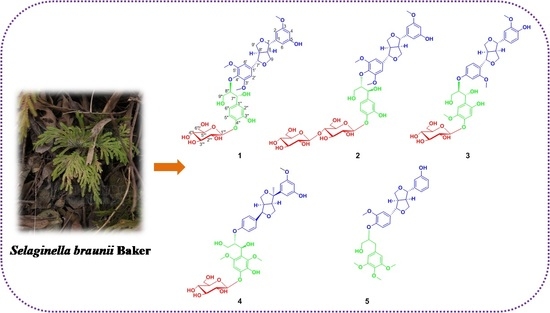
Brasesquilignan A–E, Five New Furofurans Lignans from Selaginella braunii Baker
Abstract
:1. Introduction
2. Results and Discussion
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Plant Material
3.3. Extraction and Isolation
3.4. Enzymatic Hydrolysis
3.5. Anti-Proliferative Evaluation
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Editorial Committee of the Flora of China. Flora of China; Science Press: Beijing, China, 2004; p. 108. [Google Scholar]
- Wang, Y.H.; Sun, Q.Y.; Yang, F.M.; Long, C.L.; Zhao, F.W.; Tang, G.H.; Niu, H.M.; Wang, H.; Huang, Q.Q.; Xu, J.J. Neolignans and caffeoyl derivatives from Selaginella moellendorffii. Helv. Chim. Acta 2010, 93, 2467–2477. [Google Scholar] [CrossRef]
- Cheng, F.; Xu, K.P.; Liu, L.F.; Yao, C.P.; Xu, P.S.; Zhou, G.; Li, D.; Li, X.M.; Chen, K.; Zou, Z.X.; et al. New neolignans from Selaginella picta and their protective effect on HT-22 cells. Fitoterapia 2018, 127, 69–73. [Google Scholar] [CrossRef] [PubMed]
- Long, H.P.; Li, F.S.; Xu, K.P.; Yang, Z.B.; Li, J.; Peng, J.; Tan, G.S. Bioactive compounds from Selaginella involven Spring that protect PC-12 cells. Chin. Chem. Lett. 2014, 25, 805–808. [Google Scholar] [CrossRef]
- Lin, R.C.; Skaltsounis, A.L.; Seguin, E.; Tillequin, F.; Koch, M. Phenolic constituents of Selaginella doederleinii. Planta Med. 1994, 60, 168–170. [Google Scholar] [CrossRef]
- Feng, W.S.; Chen, H.; Zheng, X.K.; Wang, Y.Z.; Gao, L.; Li, H.W. Two new secolignans from Selaginella sinensis (Desv.) Spring. J. Asian Nat. Prod. Res. 2009, 11, 658–662. [Google Scholar] [CrossRef] [PubMed]
- Zhuo, J.X.; Wang, Y.H.; Su, X.L.; Mei, R.Q.; Yang, J.; Kong, Y.; Long, C.L. Neolignans from Selaginella moellendorffii. Nat. Prod. Bioprospect. 2016, 6, 161–166. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Wang, J. Phenolic compounds from Selaginella moellendorffii. Chem.Biodivers. 2011, 8, 1735–1747. [Google Scholar] [CrossRef] [PubMed]
- Katagiri, S.; Watanabe, Y.; Yaoita, Y.; Kikuchi, M.; Machida, K. Two new phenolic glycosides from Viburnum plicatum var. plicatum f. plicatum. Nat. Prod. Commun. 2011, 6, 1901–1904. [Google Scholar] [CrossRef] [PubMed]
- Shao, S.Y.; Yang, Y.N.; Feng, Z.M.; Jiang, J.S.; Zhang, P.C. An efficient method for determining the relative configuration of furofuran lignans by 1H NMR Spectroscopy. J. Nat. Prod. 2018, 81, 1023–1028. [Google Scholar] [CrossRef] [PubMed]
- Greger, H.; Hofer, O. New unsymmetrically substituted tetrahydrofurofuran lignans from artemisia absinthium: Assignment of the relative stereochemistry by lanthanide induced chemical shifts. Tetrahedron 1980, 36, 3551–3558. [Google Scholar] [CrossRef]
- Zhao, M.Z.; Shen, Y.; Xu, W.; Chen, Y.Z.; Jiang, B. A new lignan glycoside from Astragalus yunnanensis. J. Asian Nat. Prod. Res. 2019, 22, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Park, S.Y.; Hong, S.S.; Han, X.H.; Hwang, J.S.; Lee, D.H.; Ro, J.S.; Hwang, B.Y. Lignans from Arctium lappa and their inhibition ofLPS-induced nitric oxide production. Chem. Pharm. Bull. 2007, 55, 150–152. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.H.; Moon, E.; Kim, S.Y.; Lee, K.R. Lignans from the tuber-barks of Colocasis antiquorum var. esculenta and theirantimelanogenic activity. J. Agric. Food Chem. 2010, 58, 4779–4785. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Ali, Z.; Li, X.C.; Khan, I.A. Neolignans from the leaves of Casearia sylvestris Swartz. Helv. Chim. Acta 2010, 93, 139–146. [Google Scholar] [CrossRef]
- Xiao, H.H.; Dai, Y.; Wong, M.S.; Yao, X.S. New lignans from the bioactive fraction of Sambucus williamsii Hance and proliferation activities on osteoblastic-like UMR106 cells. Fitoterapia 2014, 94, 29–35. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Zhu, C.G.; Li, Y.R.; Tian, Y.; Lin, S.; Yuan, S.P.; Hu, J.F.; Hou, Q.; Chen, N.H.; Yang, Y.C.; et al. Lignans and neolignans from Sinocalamus affinis and their absolute configurations. J. Nat. Prod. 2011, 74, 1188–1200. [Google Scholar] [CrossRef]
- Muhit, M.A.; Umehara, K.; Mori-Yasumoto, K.; Noguchi, H. Furofuran Lignan Glucosides with Estrogen-Inhibitory Properties from the Bangladeshi Medicinal Plant Terminalia citrina. J. Nat. Prod. 2016, 79, 1298–1307. [Google Scholar] [CrossRef] [PubMed]
- Greca, M.D.; Molinaro, A.; Monaco, P.; Previtera, L. Lignans from Arum italicum. Phytochemistry 1994, 35, 777–779. [Google Scholar] [CrossRef]



| Position | 1 | 2 | 3 | 4 | 5 |
|---|---|---|---|---|---|
| 1 | |||||
| 2 | 6.87, brs | 6.86, brs | 6.88, d (1.6) | 6.70, m | 6.89, d (1.4) |
| 3 | |||||
| 4 | 6.78, m | 6.78, m | 6.70, m | 6.74, d (1.6) | |
| 5 | 6.72, d (8.1) | 6.59, m | |||
| 6 | 7.03, brs | 7.03, brs | 6.75, d (1.6) | 6.82, brs | 6.76, d (1.6) |
| 7 | 4.63, d (4.1) | 4.62, d (4.0) | 4.63, t (4.2) | 4.60, m | |
| 8 | 3.06, m | 3.05, m | 3.06, m | 3.47 overlapped | 3.03, m |
| 9a | 4.13, m | 4.15, m | 4.13, m | 3.88, m | 4.13, m |
| 9b | 3.73, m | 3.72, m | 3.73, m | 3.57, m | 3.73, m |
| 1′ | |||||
| 2′ | 6.75, m | 6.73, m | 6.82, brs | 6.60, brs | |
| 3′ | 6.97, d (1.9) | 6.78, dd (1.6, 8.0) | |||
| 4′ | |||||
| 5′ | 6.85, dd (8.6, 1.8) | 6.97, d (1.5) | 6.65, d (8.0) | ||
| 6′ | 6.89, d (1.4) | 6.88, brs | 7.06, d (8.5) | 6.70, m | 6.60, brs |
| 7′ | 4.61, d (4.1) | 4.64, d (4.0) | 4.63, t (4.2) | 4.63, d (6.4) | 4.60, m |
| 8′ | 3.06, m | 3.05, m | 3.06, m | 2.19, m | 3.03, m |
| 9′a | 4.13, m | 4.15, m | 4.13, m | 3.68, m | 4.13, m |
| 9′b | 3.73, m | 3.72, m | 3.73, m | 3.46 overlapped | 3.73, m |
| 1″ | |||||
| 2″ | 6.97, d (1.6) | 6.96, brs | 6.73, brs | ||
| 3″ | 6.87, brs | ||||
| 4″ | |||||
| 5″ | 6.73, m | 6.75, m | 6.74, brs | ||
| 6″ | 6.78, m | 6.78, m | 6.86, brs | 6.71, brs | |
| 7″ | 3.61, m | 3.58, m | 3.43, m | 3.59, m | 4.11, m3.40 overlapped |
| 8″ | 5.47, d (7.4) | 5.45, d (7.4) | 5.51, d (6.5) | 5.46, d (7.2) | 4.12, m |
| 9″a | 3.97, m | 3.72, m | 3.72, m | 3.95, m | 4.11, m |
| 9″b | 3.72, m | 3.04, m | 3.63, m | 3.06, m | 3.40 overlapped |
| 1‴ | 4.25, d (7.8) | 4.24, d (7.7) | 4.89, d (7.4) | 4.23, d (7.8) | |
| 2‴ | 3.00, m | 2.97, m | 3.25, m | 3.00, m | |
| 3‴ | 3.08, m | 3.05, m | 3.25, m | 3.11, m | |
| 4‴ | 3.06, m | 3.06, m | 3.14, m | 3.06, m | |
| 5‴ | 3.18, m | 3.16, m | 3.26, m | 3.10, m | |
| 6‴ | 3.65, m 3.42, overlapped | 3.65, m 3.42, overlapped | 3.64, m 3.43, m | 3.66, m 3.43 overlapped | |
| –OCH3 | (3-) 3.79, brs | (3-) 3.78, brs | (4-) 3.77, brs | (3-) 3.76, brs | (3″-) 3.71, brs |
| (3′-) 3.76, brs | (3′-) 3.76, brs | (2′-) 3.75, brs | (2″-) 3.74, brs | (3′-) 3.74, brs | |
| (5′-) 3.76, brs | (5′-) 3.75, brs | (3″-) 3.80, brs | (6″-) 3.74, brs | (4″-) 3.75, brs | |
| (5″-) 3.76, brs | |||||
| 7-CH3 | 1.07, d (6.7) | ||||
| 1″″ | 4.25, d (7.7) | ||||
| 2″″ | 2.98, m | ||||
| 3″″ | 3.05, m | ||||
| 4″″ | 3.05, m | ||||
| 5″″ | 3.33, m | ||||
| 6″″ | 3.50 overlapped |
| Position | 1 | 2 | 3 | 4 | 5 |
|---|---|---|---|---|---|
| 1 | 135.2 | 135.2 | 132.6 | 134.5 | 145.1 |
| 2 | 111.0 | 110.9 | 110.9 | 113.2 | 110.9 |
| 3 | 144.0 | 144.0 | 148.0 | 143.9 | 146.4 |
| 4 | 119.2 | 119.2 | 146.4 | 115.5 | 119.1 |
| 5 | 147.3 | 147.3 | 115.6 | 146.0 | 122.1 |
| 6 | 115.6 | 115.6 | 119.0 | 117.6 | 114.1 |
| 7 | 85.9 | 85.9 | 85.5 | 79.6 | 85.6 |
| 8 | 54.3 | 54.2 | 54.0 | 67.5 | 54.0 |
| 9 | 71.5 | 71.4 | 71.5 | 72.3 | 71.4 |
| 1′ | 146.6 | 146.4 | 146.6 | 135.1 | 135.3 |
| 2′ | 115.6 | 115.7 | 149.5 | 110.4 | 104.1 |
| 3′ | 148.0 | 148.0 | 110.9 | 119.1 | 148.0 |
| 4′ | 132.0 | 132.1 | 135.7 | 146.1 | 129.8 |
| 5′ | 148.1 | 148.1 | 118.3 | 110.9 | 115.6 |
| 6′ | 111.0 | 110.9 | 115.6 | 118.7 | 104.1 |
| 7′ | 85.6 | 85.6 | 85.9 | 82.3 | 85.8 |
| 8′ | 54.0 | 54.0 | 54.3 | 52.9 | 54.1 |
| 9′ | 71.3 | 71.4 | 71.5 | 59.0 | 71.5 |
| 1″ | 129.5 | 129.5 | 129.4 | 129.2 | 129.1 |
| 2″ | 111.1 | 110.9 | 147.3 | 147.8 | 115.6 |
| 3″ | 147.3 | 147.3 | 115.6 | 146.8 | 148.4 |
| 4″ | 132.5 | 132.6 | 135.0 | 132.4 | 153.6 |
| 5″ | 115.7 | 115.6 | 144.0 | 115.6 | 148.4 |
| 6″ | 119.2 | 119.1 | 110.9 | 148.0 | 104.1 |
| 7″ | 51.0 | 51.0 | 53.9 | 50.9 | 61.9 |
| 8″ | 87.4 | 87.2 | 87.2 | 87.3 | 84.0 |
| 9″ | 70.6 | 70.6 | 63.4 | 70.5 | 62.2 |
| 1‴ | 103.3 | 103.2 | 100.5 | 103.2 | |
| 2‴ | 73.9 | 73.9 | 73.6 | 73.9 | |
| 3‴ | 77.3 | 77.2 | 77.3 | 77.2 | |
| 4‴ | 70.6 | 70.4 | 70.1 | 70.5 | |
| 5‴ | 77.4 | 77.3 | 77.5 | 77.4 | |
| 6‴ | 61.5 | 61.5 | 61.0 | 61.5 | |
| -OCH3 | (3-) 56.2 | (3-) 56.2 | (4-) 56.2 | (3-) 56.1 | (3″-) 56.1 |
| (3′-) 56.1 | (3′-) 56.1 | (2′-) 56.2 | (2″-) 56.0 | (3′-) 56.0 | |
| (5′-) 56.1 | (5′-) 56.1 | (3″-) 56.1 | (6″-) 56.0 | (4″-) 56.4 | |
| (5″-) 56.5 | |||||
| 7-CH3 | 22.0 | ||||
| 1″″ | 103.9 | ||||
| 2″″ | 73.8 | ||||
| 3″″ | 77.2 | ||||
| 4″″ | 68.9 | ||||
| 5″″ | 76.3 | ||||
| 6″″ | 67.5 |
| Compound | IC50 (μM) | ||||
|---|---|---|---|---|---|
| SK-MEL-28 | A375 | A549 | MCF-7 | MDA-MB-231 | |
| 1 | N/A | N/A | N/A | 93.69 ± 5.54 | N/A |
| 2 | 48.30 ± 5.29 | 35.12 ± 2.54 | 27.82 ± 2.38 | 22.09 ± 2.39 | 44.02 ± 2.32 |
| 3 | 56.82 ± 4.83 | 63.57 ± 1.49 | 38.88 ± 2.85 | 31.26 ± 1.14 | 53.56 ± 1.44 |
| 4 | N/A | N/A | N/A | N/A | N/A |
| 5 | N/A | N/A | N/A | N/A | N/A |
| STS | 0.04 ± 0.008 | 0.06 ± 0.006 | 0.4 ± 0.11 | 0.2 ± 0.04 | 0.03 ± 0.005 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheng, F.; Wu, J.; Zhang, Y.; Wang, Y.; Li, G.; Zeng, H.; He, X.; Deng, G.; Tan, J.; Long, H.; et al. Brasesquilignan A–E, Five New Furofurans Lignans from Selaginella braunii Baker. Molecules 2022, 27, 6349. https://doi.org/10.3390/molecules27196349
Cheng F, Wu J, Zhang Y, Wang Y, Li G, Zeng H, He X, Deng G, Tan J, Long H, et al. Brasesquilignan A–E, Five New Furofurans Lignans from Selaginella braunii Baker. Molecules. 2022; 27(19):6349. https://doi.org/10.3390/molecules27196349
Chicago/Turabian StyleCheng, Fei, Jianping Wu, Yan Zhang, Yuyan Wang, Guihua Li, Hongliang Zeng, Xiaoai He, Guiming Deng, Jianbin Tan, Hongping Long, and et al. 2022. "Brasesquilignan A–E, Five New Furofurans Lignans from Selaginella braunii Baker" Molecules 27, no. 19: 6349. https://doi.org/10.3390/molecules27196349
APA StyleCheng, F., Wu, J., Zhang, Y., Wang, Y., Li, G., Zeng, H., He, X., Deng, G., Tan, J., Long, H., Zeng, P., Liu, Y., Zhu, G., Chen, Z., & Xu, K. (2022). Brasesquilignan A–E, Five New Furofurans Lignans from Selaginella braunii Baker. Molecules, 27(19), 6349. https://doi.org/10.3390/molecules27196349