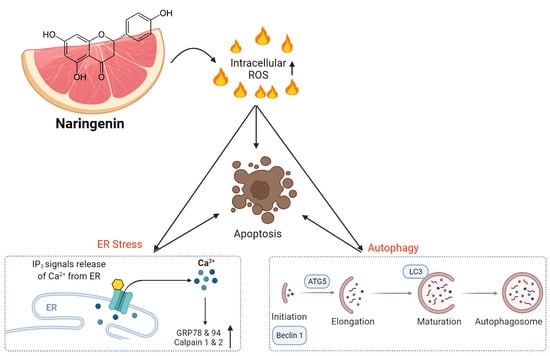
Naringenin Induces ROS-Mediated ER Stress, Autophagy, and Apoptosis in Human Osteosarcoma Cell Lines
Abstract
:1. Introduction
2. Materials and Methods
2.1. Material
2.2. Cell Line and Cell Culture
2.3. Cell Viability/Proliferation Assay
2.4. Colony Formation Assay
2.5. Cytosolic and Mitochondrial Protein Extraction
2.6. Western Blot Analysis
2.7. DAPI Staining
2.8. Terminal Deoxynucleotidyl Transferase-Mediated dUTP Nick-End Labeling Assay
2.9. Annexin V and Propidium Iodide Staining
2.10. Cell Cycle Analysis Using PI Staining
2.11. Intracellular ROS Production, Ca2+ Concentration, and Mitochondrial Mmembrane Potential
2.12. Small Interfering RNA Transfection
2.13. Autophagy Assay
2.14. Caspase Activity Assay
2.15. Transmission Electron Microscopy
2.16. Statistical Analysis
3. Results
3.1. Induction of Cell Apoptosis in Human Osteosarcoma Cells by Naringenin
3.2. Induction of ROS-Mediated ER Stress by Naringenin in Osteosarcoma Cells
3.3. Involement of Cell Apoptosis in Naringenin-Induced Mitochondrial Dysfunction in Human Osteosarcoma
3.4. Triggering of Autophagy in Human Osteosarcoma Cells by Naringenin
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Li, T.; Jiang, S.; Yang, Y. Database Selection and Heterogeneity-More Details, More Credibility. JAMA Oncol. 2018, 4, 1295. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Mirabello, L.; Troisi, R.J.; Savage, S.A. Osteosarcoma incidence and survival rates from 1973 to 2004: Data from the Surveillance, Epidemiology, and End Results Program. Cancer 2009, 115, 1531–1543. [Google Scholar] [CrossRef] [Green Version]
- Whelan, J.S.; Jinks, R.C.; McTiernan, A.; Sydes, M.R.; Hook, J.M.; Trani, L.; Uscinska, B.; Bramwell, V.; Lewis, I.J.; Nooij, M.A.; et al. Survival from high-grade localised extremity osteosarcoma: Combined results and prognostic factors from three European Osteosarcoma Intergroup randomised controlled trials. Ann. Oncol. 2012, 23, 1607–1616. [Google Scholar] [CrossRef] [PubMed]
- Bielack, S.S.; Kempf-Bielack, B.; Delling, G.; Exner, G.U.; Flege, S.; Helmke, K.; Kotz, R.; Salzer-Kuntschik, M.; Werner, M.; Winkelmann, W.; et al. Prognostic factors in high-grade osteosarcoma of the extremities or trunk: An analysis of 1702 patients treated on neoadjuvant cooperative osteosarcoma study group protocols. J. Clin. Oncol. 2002, 20, 776–790. [Google Scholar] [CrossRef]
- Jawad, M.U.; Cheung, M.C.; Clarke, J.; Koniaris, L.G.; Scully, S.P. Osteosarcoma: Improvement in survival limited to high-grade patients only. J. Cancer Res. Clin. Oncol. 2011, 137, 597–607. [Google Scholar] [CrossRef] [PubMed]
- Jiang, F.; Shi, Y.; Li, G.J.; Zhou, F. A meta-analysis of limb-salvage versus amputation in the treatment of patients with Ennekingdouble daggerU pathologic fracture osteosarcoma. Indian J. Cancer 2015, 51 (Suppl. 2), e21–e24. [Google Scholar] [CrossRef]
- Gill, J.; Gorlick, R. Advancing therapy for osteosarcoma. Nat. Rev. Clin. Oncol. 2021, 18, 609–624. [Google Scholar] [CrossRef]
- Naveen Kumar, D.R.; Cijo George, V.; Suresh, P.K.; Ashok Kumar, R. Cytotoxicity, apoptosis induction and anti-metastatic potential of Oroxylum indicum in human breast cancer cells. Asian Pac. J. Cancer Prev. 2012, 13, 2729–2734. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cragg, G.M.; Newman, D.J. Plants as a source of anti-cancer agents. J. Ethnopharmacol. 2005, 100, 72–79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yanez, J.A.; Andrews, P.K.; Davies, N.M. Methods of analysis and separation of chiral flavonoids. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2007, 848, 159–181. [Google Scholar] [CrossRef]
- Mandalari, G.; Bennett, R.N.; Bisignano, G.; Trombetta, D.; Saija, A.; Faulds, C.B.; Gasson, M.J.; Narbad, A. Antimicrobial activity of flavonoids extracted from bergamot (Citrus bergamia Risso) peel, a byproduct of the essential oil industry. J. Appl. Microbiol. 2007, 103, 2056–2064. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, R.J.; Baptista, R.; Moreno, A.; Madeira, P.G.; Khonkarn, R.; Baubichon-Cortay, H.; Dos Santos, D.J.; Falson, P.; Ferreira, M.U. Optimizing the flavanone core toward new selective nitrogen-containing modulators of ABC transporters. Future Med. Chem. 2018, 10, 725–741. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Tiku, A.B. Biochemical and Molecular Mechanisms of Radioprotective Effects of Naringenin, a Phytochemical from Citrus Fruits. J. Agric. Food Chem. 2016, 64, 1676–1685. [Google Scholar] [CrossRef]
- Kanno, S.; Tomizawa, A.; Hiura, T.; Osanai, Y.; Shouji, A.; Ujibe, M.; Ohtake, T.; Kimura, K.; Ishikawa, M. Inhibitory effects of naringenin on tumor growth in human cancer cell lines and sarcoma S-180-implanted mice. Biol. Pharm. Bull 2005, 28, 527–530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Z.; Jin, G.; Ge, Y.; Guo, Z. Naringenin inhibits migration of breast cancer cells via inflammatory and apoptosis cell signaling pathways. Inflammopharmacology 2019, 27, 1021–1036. [Google Scholar] [CrossRef] [PubMed]
- Song, H.M.; Park, G.H.; Eo, H.J.; Jeong, J.B. Naringenin-Mediated ATF3 Expression Contributes to Apoptosis in Human Colon Cancer. Biomol. Ther. 2016, 24, 140–146. [Google Scholar] [CrossRef] [Green Version]
- Jin, C.Y.; Park, C.; Hwang, H.J.; Kim, G.Y.; Choi, B.T.; Kim, W.J.; Choi, Y.H. Naringenin up-regulates the expression of death receptor 5 and enhances TRAIL-induced apoptosis in human lung cancer A549 cells. Mol. Nutr. Food Res. 2011, 55, 300–309. [Google Scholar] [CrossRef]
- Camuzard, O.; Santucci-Darmanin, S.; Carle, G.F.; Pierrefite-Carle, V. Role of autophagy in osteosarcoma. J. Bone Oncol. 2019, 16, 100235. [Google Scholar] [CrossRef] [PubMed]
- Niu, J.; Yan, T.; Guo, W.; Wang, W.; Zhao, Z. Insight into the Role of Autophagy in Osteosarcoma and Its Therapeutic Implication. Front. Oncol. 2019, 9, 1232. [Google Scholar] [CrossRef]
- Yoon, S.Y.; Kim, D.H. Alzheimer’s disease genes and autophagy. Brain Res. 2016, 1649, 201–209. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Meng, Y.; Zong, C.; Zhang, S.; Wei, L. Autophagy and Tumorigenesis. Adv. Exp. Med. Biol. 2020, 1207, 275–299. [Google Scholar] [CrossRef]
- Liao, Y.X.; Yu, H.Y.; Lv, J.Y.; Cai, Y.R.; Liu, F.; He, Z.M.; He, S.S. Targeting autophagy is a promising therapeutic strategy to overcome chemoresistance and reduce metastasis in osteosarcoma. Int. J. Oncol. 2019, 55, 1213–1222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, L.; Loveless, J.; Shay, C.; Teng, Y. Targeting ROS-Mediated Crosstalk Between Autophagy and Apoptosis in Cancer. Adv. Exp. Med. Biol. 2020, 1260, 1–12. [Google Scholar] [CrossRef]
- Bhandary, B.; Marahatta, A.; Kim, H.R.; Chae, H.J. An involvement of oxidative stress in endoplasmic reticulum stress and its associated diseases. Int. J. Mol. Sci. 2012, 14, 434–456. [Google Scholar] [CrossRef] [PubMed]
- Tian, J.; Lu, Z.; Wang, Y.; Zhang, M.; Wang, X.; Tang, X.; Peng, X.; Zeng, H. Nerol triggers mitochondrial dysfunction and disruption via elevation of Ca2+ and ROS in Candida albicans. Int. J. Biochem. Cell Biol. 2017, 85, 114–122. [Google Scholar] [CrossRef]
- Lu, Y.; Zhang, C.; Bucheli, P.; Wei, D. Citrus flavonoids in fruit and traditional Chinese medicinal food ingredients in China. Plant Foods Hum. Nutr. 2006, 61, 57–65. [Google Scholar] [CrossRef]
- Miyata, M.; Takano, H.; Takahashi, K.; Sasaki, Y.F.; Yamazoe, Y. Suppression of 2-amino-1-methyl-6-phenylimidazo [4,5-b]pyridine-induced DNA damage in rat colon after grapefruit juice intake. Cancer Lett. 2002, 183, 17–22. [Google Scholar] [CrossRef]
- Zobeiri, M.; Belwal, T.; Parvizi, F.; Naseri, R.; Farzaei, M.H.; Nabavi, S.F.; Sureda, A.; Nabavi, S.M. Naringenin and its Nano-formulations for Fatty Liver: Cellular Modes of Action and Clinical Perspective. Curr. Pharm. Biotechnol. 2018, 19, 196–205. [Google Scholar] [CrossRef]
- Chandrika, B.B.; Steephan, M.; Kumar, T.R.S.; Sabu, A.; Haridas, M. Hesperetin and Naringenin sensitize HER2 positive cancer cells to death by serving as HER2 Tyrosine Kinase inhibitors. Life Sci. 2016, 160, 47–56. [Google Scholar] [CrossRef]
- Arul, D.; Subramanian, P. Naringenin (citrus flavonone) induces growth inhibition, cell cycle arrest and apoptosis in human hepatocellular carcinoma cells. Pathol. Oncol. Res. 2013, 19, 763–770. [Google Scholar] [CrossRef]
- Guo, S.; Pridham, K.J.; Virbasius, C.M.; He, B.; Zhang, L.; Varmark, H.; Green, M.R.; Sheng, Z. A large-scale RNA interference screen identifies genes that regulate autophagy at different stages. Sci. Rep. 2018, 8, 2822. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, W.P.; Klionsky, D.J. Autophagy in yeast: A review of the molecular machinery. Cell Struct. Funct. 2002, 27, 409–420. [Google Scholar] [CrossRef] [Green Version]
- Hasima, N.; Ozpolat, B. Regulation of autophagy by polyphenolic compounds as a potential therapeutic strategy for cancer. Cell Death Dis. 2014, 5, e1509. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiong, X.; Wu, M.; Zhang, H.; Li, J.; Lu, B.; Guo, Y.; Zhou, T.; Guo, H.; Peng, R.; Li, X.; et al. Atg5 siRNA inhibits autophagy and enhances norcantharidin-induced apoptosis in hepatocellular carcinoma. Int. J. Oncol. 2015, 47, 1321–1328. [Google Scholar] [CrossRef] [Green Version]
- Graf, M.R.; Jia, W.; Johnson, R.S.; Dent, P.; Mitchell, C.; Loria, R.M. Autophagy and the functional roles of Atg5 and beclin-1 in the anti-tumor effects of 3beta androstene 17alpha diol neuro-steroid on malignant glioma cells. J. Steroid Biochem. Mol. Biol. 2009, 115, 137–145. [Google Scholar] [CrossRef] [PubMed]
- Dai, G.; Zheng, D.; Guo, W.; Yang, J.; Cheng, A.Y. Cinobufagin Induces Apoptosis in Osteosarcoma Cells Via the Mitochondria-Mediated Apoptotic Pathway. Cell Physiol. Biochem. 2018, 46, 1134–1147. [Google Scholar] [CrossRef]
- Yu, X.; Zhou, X.; Fu, C.; Wang, Q.; Nie, T.; Zou, F.; Guo, R.; Liu, H.; Zhang, B.; Dai, M. Celastrol induces apoptosis of human osteosarcoma cells via the mitochondrial apoptotic pathway. Oncol. Rep. 2015, 34, 1129–1136. [Google Scholar] [CrossRef]
- Sauer, H.; Wartenberg, M.; Hescheler, J. Reactive oxygen species as intracellular messengers during cell growth and differentiation. Cell Physiol. Biochem. 2001, 11, 173–186. [Google Scholar] [CrossRef]
- Bossis, G.; Sarry, J.E.; Kifagi, C.; Ristic, M.; Saland, E.; Vergez, F.; Salem, T.; Boutzen, H.; Baik, H.; Brockly, F.; et al. The ROS/SUMO axis contributes to the response of acute myeloid leukemia cells to chemotherapeutic drugs. Cell Rep. 2014, 7, 1815–1823. [Google Scholar] [CrossRef] [Green Version]
- Ivanova, D.; Zhelev, Z.; Aoki, I.; Bakalova, R.; Higashi, T. Overproduction of reactive oxygen species—Obligatory or not for induction of apoptosis by anticancer drugs. Chin. J. Cancer Res. 2016, 28, 383–396. [Google Scholar] [CrossRef] [Green Version]
- Magnano, S.; Hannon Barroeta, P.; Duffy, R.; O’Sullivan, J.; Zisterer, D.M. Cisplatin induces autophagy-associated apoptosis in human oral squamous cell carcinoma (OSCC) mediated in part through reactive oxygen species. Toxicol. Appl. Pharmacol. 2021, 427, 115646. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Zhu, Y.; Yu, H.; Liu, X.; Jiao, B.; Lu, X. Libertellenone H, a Natural Pimarane Diterpenoid, Inhibits Thioredoxin System and Induces ROS-Mediated Apoptosis in Human Pancreatic Cancer Cells. Molecules 2021, 26, 315. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Xu, Y.; Sun, J.; Sui, Z. The Novel Curcumin Derivative 1g Induces Mitochondrial and ER-Stress-Dependent Apoptosis in Colon Cancer Cells by Induction of ROS Production. Front. Oncol. 2021, 11, 644197. [Google Scholar] [CrossRef]
- Park, H.J.; Choi, Y.J.; Lee, J.H.; Nam, M.J. Naringenin causes ASK1-induced apoptosis via reactive oxygen species in human pancreatic cancer cells. Food Chem. Toxicol. 2017, 99, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Datta, K.; Babbar, P.; Srivastava, T.; Sinha, S.; Chattopadhyay, P. p53 dependent apoptosis in glioma cell lines in response to hydrogen peroxide induced oxidative stress. Int. J. Biochem. Cell Biol. 2002, 34, 148–157. [Google Scholar] [CrossRef]
- Zhang, Y.; He, N.; Zhou, X.; Wang, F.; Cai, H.; Huang, S.H.; Chen, X.; Hu, Z.; Jin, X. Betulinic acid induces autophagy-dependent apoptosis via Bmi-1/ROS/AMPK-mTOR-ULK1 axis in human bladder cancer cells. Aging 2021, 13, 21251–21267. [Google Scholar] [CrossRef]
- Xipell, E.; Gonzalez-Huarriz, M.; Martinez de Irujo, J.J.; Garcia-Garzon, A.; Lang, F.F.; Jiang, H.; Fueyo, J.; Gomez-Manzano, C.; Alonso, M.M. Salinomycin induced ROS results in abortive autophagy and leads to regulated necrosis in glioblastoma. Oncotarget 2016, 7, 30626–30641. [Google Scholar] [CrossRef]
- Yoon, M.J.; Kang, Y.J.; Kim, I.Y.; Kim, E.H.; Lee, J.A.; Lim, J.H.; Kwon, T.K.; Choi, K.S. Monensin, a polyether ionophore antibiotic, overcomes TRAIL resistance in glioma cells via endoplasmic reticulum stress, DR5 upregulation and c-FLIP downregulation. Carcinogenesis 2013, 34, 1918–1928. [Google Scholar] [CrossRef] [Green Version]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, C.-W.; Huang, C.C.-Y.; Chi, M.-C.; Lee, K.-H.; Peng, K.-T.; Fang, M.-L.; Chiang, Y.-C.; Liu, J.-F. Naringenin Induces ROS-Mediated ER Stress, Autophagy, and Apoptosis in Human Osteosarcoma Cell Lines. Molecules 2022, 27, 373. https://doi.org/10.3390/molecules27020373
Lee C-W, Huang CC-Y, Chi M-C, Lee K-H, Peng K-T, Fang M-L, Chiang Y-C, Liu J-F. Naringenin Induces ROS-Mediated ER Stress, Autophagy, and Apoptosis in Human Osteosarcoma Cell Lines. Molecules. 2022; 27(2):373. https://doi.org/10.3390/molecules27020373
Chicago/Turabian StyleLee, Chiang-Wen, Cathy Chia-Yu Huang, Miao-Ching Chi, Kuan-Han Lee, Kuo-Ti Peng, Mei-Ling Fang, Yao-Chang Chiang, and Ju-Fang Liu. 2022. "Naringenin Induces ROS-Mediated ER Stress, Autophagy, and Apoptosis in Human Osteosarcoma Cell Lines" Molecules 27, no. 2: 373. https://doi.org/10.3390/molecules27020373
APA StyleLee, C. -W., Huang, C. C. -Y., Chi, M. -C., Lee, K. -H., Peng, K. -T., Fang, M. -L., Chiang, Y. -C., & Liu, J. -F. (2022). Naringenin Induces ROS-Mediated ER Stress, Autophagy, and Apoptosis in Human Osteosarcoma Cell Lines. Molecules, 27(2), 373. https://doi.org/10.3390/molecules27020373