Cytotoxic Activity and Docking Studies of 2-arenoxybenzaldehyde N-acyl Hydrazone and 1,3,4-Oxadiazole Derivatives against Various Cancer Cell Lines
Abstract
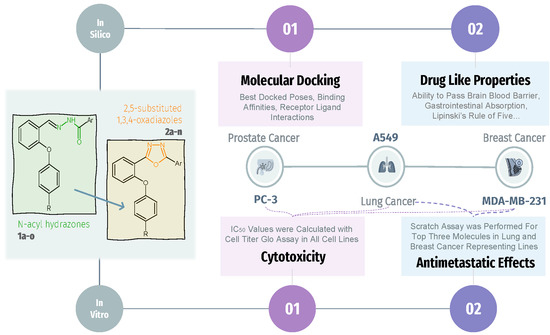
:1. Introduction
2. Results
2.1. Biologic Activities
2.1.1. In Vitro Cell Viability Assays
2.1.2. Scratch Assay
2.2. Molecular Docking Studies
2.3. Drug-like Properties
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Cell Viability Assay
4.3. Scratch Assay
4.4. Statistical Analysis
4.5. Molecular Docking Studies and Drug-like Properties
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Siegel, R.; Miller, K.; Fuchs, H.; Jemal, A. Cancer Statistics, 2022. CA A Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Peng, J.; Zhou, W.; Qiao, H.; Deng, X.; Li, Z.; Li, J.; Fu, Y.; Li, S.; Sun, K.; et al. Potent Hydrazone Derivatives Targeting Esophageal Cancer Cells. Eur. J. Med. Chem. 2018, 148, 359–371. [Google Scholar] [CrossRef] [PubMed]
- Halil, Ş.; Berre, M.; Rabia Büşra, Ş.; Halil Burak, K.; Ebru, H. Synthesis Of Oleanolic Acid Hydrazide-Hydrazone Hybrid Derivatives And Investigation Of Their Cytotoxic Effects On A549 Human Lung Cancer Cells. Results Chem. 2022, 4, 100317. [Google Scholar] [CrossRef]
- Iliev, I.; Kontrec, D.; Detcheva, R.; Georgieva, M.; Balacheva, A.; Galić, N.; Pajpanova, T. Cancer Cell Growth Inhibition By Aroylhydrazone Derivatives. Biotechnol. Biotechnol. Equip. 2019, 33, 756–763. [Google Scholar] [CrossRef] [Green Version]
- Świątek, P.; Saczko, J.; Rembiałkowska, N.; Kulbacka, J. Synthesis Of New Hydrazone Derivatives And Evaluation Of Their Efficacy As Proliferation Inhibitors In Human Cancer Cells. Med. Chem. 2019, 15, 903–910. [Google Scholar] [CrossRef] [PubMed]
- Popiołek, Ł. Hydrazide–Hydrazones as Potential Antimicrobial Agents: Overview of The Literature Since 2010. Med. Chem. Res. 2016, 26, 287–301. [Google Scholar] [CrossRef] [Green Version]
- Pieczonka, A.; Strzelczyk, A.; Sadowska, B.; Mlostoń, G.; Stączek, P. Synthesis and Evaluation of Antimicrobial Activity of Hydrazones Derived from 3-Oxido-1H-Imidazole-4-Carbohydrazides. Eur. J. Med. Chem. 2013, 64, 389–395. [Google Scholar] [CrossRef]
- Kajal, A.; Bala, S.; Sharma, N.; Kamboj, S.; Saini, V. Therapeutic Potential of Hydrazones as Anti-Inflammatory Agents. Int. J. Med. Chem. 2014, 2014, 76103. [Google Scholar] [CrossRef]
- Alam, M.; Verma, G.; Shaquiquzzaman, M.; Marella, A.; Akhtar, M.; Ali, M. A Review Exploring Biological Activities Of Hydrazones. J. Pharm. Bioallied Sci. 2014, 6, 69. [Google Scholar] [CrossRef]
- El-Tombary, A.; El-Hawash, S. Synthesis, Antioxidant, Anticancer And Antiviral Activities Of Novel Quinoxaline Hydrazone Derivatives And Their Acyclic C-Nucleosides. Med. Chem. 2014, 10, 521–532. [Google Scholar] [CrossRef]
- Tian, J.; Ji, R.; Wang, H.; Li, S.; Zhang, G. Discovery Of Novel A-Aminophosphonates With Hydrazone As Potential Antiviral Agents Combined With Active Fragment And Molecular Docking. Front. Chem. 2022, 10, 911453. [Google Scholar] [CrossRef]
- Glomb, T.; Świątek, P. Antimicrobial Activity Of 1,3,4-Oxadiazole Derivatives. Int. J. Mol. Sci. 2021, 22, 6979. [Google Scholar] [CrossRef] [PubMed]
- Tresse, C.; Radigue, R.; Gomes Von Borowski, R.; Thepaut, M.; Hanh Le, H.; Demay, F.; Georgeault, S.; Dhalluin, A.; Trautwetter, A.; Ermel, G.; et al. Synthesis And Evaluation Of 1,3,4-Oxadiazole Derivatives For Development As Broad-Spectrum Antibiotics. Bioorg. Med. Chem. 2019, 27, 115097. [Google Scholar] [CrossRef] [PubMed]
- Chawla, G.; Naaz, B.; Siddiqui, A. Exploring 1,3,4-Oxadiazole Scaffold For Anti-Inflammatory And Analgesic Activities: A Review Of Literature From 2005-2016. Mini-Rev. Med. Chem. 2018, 18, 216–233. [Google Scholar] [CrossRef] [PubMed]
- Alisi, I.; Uzairu, A.; Abechi, S. Free Radical Scavenging Mechanism Of 1,3,4-Oxadiazole Derivatives: Thermodynamics Of O–H And N–H Bond Cleavage. Heliyon 2020, 6, 3683. [Google Scholar] [CrossRef] [PubMed]
- Gümrükçüoğlu, N.; Bilgin Sökmen, B. Synthesis and Antioxidant Activities of New 2-(4-Methylphenylsulphonyl)-5Aryll,3,4-Oxadiazole Compounds. Erzincan Univ. J. Sci. Technol. 2021, 14, 232–240. [Google Scholar] [CrossRef]
- Glomb, T.; Szymankiewicz, K.; Świątek, P. Anti-Cancer Activity Of Derivatives Of 1,3,4-Oxadiazole. Molecules 2018, 23, 3361. [Google Scholar] [CrossRef] [Green Version]
- Almasirad, A.; Shafiee, A.; Abdollahi, M.; Noeparast, A.; Shahrokhinejad, N.; Vousooghi, N.; Tabatabai, S.; Khorasani, R. Synthesis And Analgesic Activity Of New 1,3,4-Oxadiazoles And 1,2,4-Triazoles. Med. Chem. Res. 2010, 20, 435–442. [Google Scholar] [CrossRef]
- Peng, F.; Liu, T.; Wang, Q.; Liu, F.; Cao, X.; Yang, J.; Liu, L.; Xie, C.; Xue, W. Antibacterial And Antiviral Activities Of 1,3,4-Oxadiazole Thioether 4H-Chromen-4-One Derivatives. J. Agric. Food Chem. 2021, 69, 11085–11094. [Google Scholar] [CrossRef]
- Li, Z.; Zhan, P.; Liu, X. 1,3,4-Oxadiazole: A Privileged Structure In Antiviral Agents. Mini-Rev. Med. Chem. 2011, 11, 1130–1142. [Google Scholar] [CrossRef]
- Noma, S.; Erzengin, M.; Tunç, T.; Balcıoğlu, S. Synthesis, Characterization And Biological Assessment Of A Novel Hydrazone As Potential Anticancer Agent And Enzyme Inhibitor. J. Mol. Struct. 2020, 1205, 127550. [Google Scholar] [CrossRef]
- Liu, T.; Sun, C.; Xing, X.; Jing, L.; Tan, R.; Luo, Y.; Huang, W.; Song, H.; Li, Z.; Zhao, Y. Synthesis And Evaluation Of 2-[2-(Phenylthiomethyl)-1H-Benzo[D] Imidazol-1-Yl) Acetohydrazide Derivatives As Antitumor Agents. Bioorg. Med. Chem. Lett. 2012, 22, 3122–3125. [Google Scholar] [CrossRef]
- Cui, Z.; Li, Y.; Ling, Y.; Huang, J.; Cui, J.; Wang, R.; Yang, X. New Class Of Potent Antitumor Acylhydrazone Derivatives Containing Furan. Eur. J. Med. Chem. 2010, 45, 5576–5584. [Google Scholar] [CrossRef]
- Tantak, M.; Klingler, L.; Arun, V.; Kumar, A.; Sadana, R.; Kumar, D. Design And Synthesis Of Bis(Indolyl)Ketohydrazide-Hydrazones: Identification Of Potent And Selective Novel Tubulin Inhibitors. Eur. J. Med. Chem. 2017, 136, 184–194. [Google Scholar] [CrossRef]
- Das Mukherjee, D.; Kumar, N.; Tantak, M.; Das, A.; Ganguli, A.; Datta, S.; Kumar, D.; Chakrabarti, G. Development Of Novel Bis (Indolyl)-Hydrazide–Hydrazone Derivatives As Potent Microtubule-Targeting Cytotoxic Agents Against A549 Lung Cancer Cells. Biochemistry 2016, 55, 3020–3035. [Google Scholar] [CrossRef]
- Nikolova-Mladenova, B.; Momekov, G.; Ivanov, D.; Bakalova, A. Design And Drug-Like Properties Of New 5-Methoxysalicylaldehyde Based Hydrazones With Anti-Breast Cancer Activity. J. Appl. Biomed. 2017, 15, 233–240. [Google Scholar] [CrossRef]
- Szczepankiewicz, B.; Liu, G.; Jae, H.; Tasker, A.; Gunawardana, I.; von Geldern, T.; Gwaltney, S.; Wu-Wong, J.; Gehrke, L.; Chiou, W.; et al. New Antimitotic Agents With Activity In Multi-Drug-Resistant Cell Lines And In Vivo Efficacy In Murine Tumor Models. J. Med. Chem. 2001, 44, 4416–4430. [Google Scholar] [CrossRef]
- Aboraia, A.; Abdel-Rahman, H.; Mahfouz, N.; EL-Gendy, M. Novel 5-(2-Hydroxyphenyl)-3-Substituted-2,3-Dihydro-1,3,4-Oxadiazole-2-Thione Derivatives: Promising Anticancer Agents. Bioorg. Med. Chem. 2006, 14, 1236–1246. [Google Scholar] [CrossRef]
- Kumar, D.; Sundaree, S.; Johnson, E.; Shah, K. An Efficient Synthesis And Biological Study Of Novel Indolyl-1,3,4-Oxadiazoles As Potent Anticancer Agents. Bioorg. Med. Chem. Lett. 2009, 19, 4492–4494. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Li, M.; Qian, S.; Guo, F.; Dang, X.; Wang, X.; Xue, Y.; Zhu, H. Synthesis And Antitumor Activity Of 1,3,4-Oxadiazole Possessing 1,4-Benzodioxan Moiety As A Novel Class Of Potent Methionine Aminopeptidase Type II Inhibitors. Bioorg. Med. Chem. Lett. 2013, 23, 2876–2879. [Google Scholar] [CrossRef] [PubMed]
- Puthiyapurayil, P.; Poojary, B.; Chikkanna, C.; Buridipad, S. Design, Synthesis And Biological Evaluation Of A Novel Series Of 1,3,4-Oxadiazole Bearing N-Methyl-4-(Trifluoromethyl)Phenyl Pyrazole Moiety As Cytotoxic Agents. Eur. J. Med. Chem. 2012, 53, 203–210. [Google Scholar] [CrossRef]
- Ren, J.; Wu, L.; Xin, W.; Chen, X.; Hu, K. Synthesis and Biological Evaluation Of Novel 4Β-(1,3,4-Oxadiazole-2-Amino)-Podophyllotoxin Derivatives. Bioorg. Med. Chem. Lett. 2012, 22, 4778–4782. [Google Scholar] [CrossRef] [PubMed]
- Küçük, H.B.; Alhonaish, A.; Yıldız, T.; Güzel, M. An Efficient Approach To Access 2,5-Disubstituted 1,3,4-Oxadiazoles By Oxidation Of 2-Arenoxybenzaldehyde N-Acyl Hydrazones With Molecular Iodine. ChemistrySelect 2022, 7, e202201391. [Google Scholar] [CrossRef]
- Lee, A.; Ramanujulu, P.; Poulsen, A.; Williams, M.; Blanchard, S.; Ma, D.; Bonday, Z.; Goh, K.; Goh, K.; Goh, M.; et al. Thieno [3,2-D]Pyrimidin-4(3H)-One Derivatives As PDK1 Inhibitors Discovered By Fragment-Based Screening. Bioorg. Med. Chem. Lett. 2012, 22, 4023–4027. [Google Scholar] [CrossRef]
- Mahendran, P.; Jeya Rajendran, A.; Balachandran, C.; Stalin, A.; Rangan, S.; Kothandapani, L.; Chennakesava Rao, K.; Awale, S.; Hiteshkumar, B. Synthesis Of Novel Β-Amino Alcohols From Phenylacetylcarbinol: Cytotoxicity Activity Against A549 Cells And Molecular Docking. Res. Chem. Intermed. 2017, 44, 535–552. [Google Scholar] [CrossRef]
- Abu Bakar, A.; Akhtar, M.; Mohd Ali, N.; Yeap, S.; Quah, C.; Loh, W.; Alitheen, N.; Zareen, S.; Ul-Haq, Z.; Shah, S. Design, Synthesis And Docking Studies Of Flavokawain B Type Chalcones And Their Cytotoxic Effects On MCF-7 And MDA-MB-231 Cell Lines. Molecules 2018, 23, 616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daina, A.; Michielin, O.; Zoete, V. Swissadme: A Free Web Tool to Evaluate Pharmacokinetics, Drug-Likeness And Medicinal Chemistry Friendliness Of Small Molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef] [Green Version]
- Berman, H. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, Q.; Li, D.; Pi, Y.; Li, J.; Sun, J.; Fang, F.; Zhong, W.; Gong, H.; Zhu, H. Novel 1,3,4-Oxadiazole Thioether Derivatives Targeting Thymidylate Synthase As Dual Anticancer/Antimicrobial Agents. Bioorg. Med. Chem. 2013, 21, 2286–2297. [Google Scholar] [CrossRef] [PubMed]
- Ashok, D.; Vanaja, B. Synthesis And Anticancer Activity Evaluation Of (E)-3-{[5-(Aryl)-1,3,4-Oxadiazol-2-Yl]Methyl}-5-(3,4,5-Trimethoxybenzylidene)Thiazolidine-2,4-Diones. Russ. J. Gen. Chem. 2016, 86, 681–685. [Google Scholar] [CrossRef]
- Polothi, R.; Raolji, G.; Kuchibhotla, V.; Sheelam, K.; Tuniki, B.; Thodupunuri, P. Synthesis And Biological Evaluation Of 1,2,4-Oxadiazole Linked 1,3,4-Oxadiazole Derivatives As Tubulin Binding Agents. Synth. Commun. 2019, 49, 1603–1612. [Google Scholar] [CrossRef]












| Molecules * | A549 | MDA-MB-231 | PC-3 | MRC-5 |
|---|---|---|---|---|
| 1a | 28.65 | 53.02 | 47.75 | 16.89 |
| 2a | 29.28 | 24.02 | 65.96 | 74.45 |
| 1b | 72.81 | 137.50 | 25.27 | 24.44 |
| 2b | 152.20 | 46.92 | 82.23 | 36.02 |
| 1c | 207.50 | 50.84 | 86.02 | 43.46 |
| 2c | 109.40 | 42.62 | 120.90 | 12.82 |
| 1d | 49.79 | 31.46 | 9.39 | 44.66 |
| 2d | 102.10 | 78.60 | 54.31 | 40.07 |
| 1e | 13.39 | 108.30 | 18.09 | 86.96 |
| 1f | 36.90 | 77.20 | 65.91 | 56.82 |
| 2f | 45.79 | 32.66 | 34.16 | 141.10 |
| 1g | 31.84 | 45.08 | 66.53 | 62.75 |
| 2g | 64.78 | 55.74 | 126.80 | 16.00 |
| 1h | 73.04 | 65.80 | 78.34 | 66.87 |
| 2h | 78.58 | 38.20 | 78.22 | 23.07 |
| 1i | 208.50 | 20.70 | 84.56 | 31.66 |
| 2i | 72.85 | 23.07 | 41.37 | 23.07 |
| 1j | 26.56 | 25.90 | 18.39 | 37.06 |
| 1k | 121.90 | 32.69 | 46.35 | 66.87 |
| 2k | 89.98 | 79.20 | 65.83 | 58.34 |
| 1l | 39.43 | 77.70 | 30.20 | 14.55 |
| 2l | 36.26 | 22.73 | 38.42 | 51.87 |
| 1m | 71.77 | 55.51 | 38.12 | 37.93 |
| 2m | 137.47 | 108.50 | 124.10 | 124.10 |
| 1n | 39.36 | 37.85 | 30.39 | 50.55 |
| 2n | 28.84 | 30.37 | 25.60 | 35.59 |
| 1o | 89.65 | 100.40 | 27.03 | 7.78 |
| Comp No. | Structures | Docked Amino Acid Residues (vdW Interactions) | Energy Score | RMSD Value | H Bond (Distance Å) |
|---|---|---|---|---|---|
| 1a |  | GLY89, ALA162, LEU212 | −9.31 | 0.96 | O of Carbonyl with H of NH of TYR161 (1.772) |
| 1b |  | TYR161, ALA162, GLY165 | −9.66 | 1.00 | None |
| 1c |  | VAL96, ALA109, ALA162 | −9.47 | 1.62 | H of NH with O of Carbonyl of GLU166 (2.115) |
| 1d |  | LEU159, SER160, LEU212 | −9.46 | 0.58 | O of Carbonyl with H of NH of ALA162 (2.241) |
| 1e |  | VAL96, ALA109, SER160 | −8.89 | 0.33 | None |
| 1f |  | VAL96, ALA109, SER160 | −9.63 | 1.50 | H of NH with O of Carbonyl of GLU166 (2.149) |
| 1g |  | LYS86, ALA162, GLU166 | −8.92 | 1.89 | None |
| 1h |  | LEU88, GLU166, LEU212 | −8.97 | 1.64 | None |
| 1i |  | VAL96, VAL143, THR222 | −10.50 | 0.50 | H of OH with O of Carbonyl of SER160 (2.064) |
| 1j |  | LEU88, LYS111, GLU166 | −9.87 | 0.38 | H of imine with H of OH of THR222 (2.230) |
| 1k |  | ALA109, ALA162, THR222 | −9.65 | 1.86 | None |
| 1l |  | VAL96, ALA109, LEU212 | −9.64 | 1.19 | H of NH with O of Carbonyl of GLU166 (2.207) |
| 1m |  | ALA109, ALA162, THR222 | −9.54 | 1.99 | H of NH with O of Carbonyl of GLU166 (2.168) |
| 1n |  | GLY89, LYS111, SER160, | −9.76 | 0.59 | O of Carbonyl with H of NH of ALA162 (2.220) |
| 1o |  | LEU88, GLY165, GLU166 | −9.61 | 0.93 | H of NH with O of Carbonyl of GLU209 (2.072) |
| 2a |  | VAL96, LEU159, THR222 | −10.47 | 0.12 | N of Oxazole with H of NH of TYR161 (2.023) |
| 2b |  | LEU159, ALA162, THR222 | 10.78 | 0.15 | N of Oxazole with H of NH of TYR161 (1.933) |
| 2c |  | VAL96, LEU159, THR222 | −10.45 | 0.86 | N of Oxazole with H of NH of ALA162 (1.912) |
| 2d |  | VAL96, LEU159, THR222 | −10.31 | 0.17 | N of Oxazole with H of NH of ALA162 (2.249) |
| 2f |  | VAL96, LEU159, THR222 | −11.13 | 0.23 | N of Oxazole with H of NH of ALA162 (1.964) |
| 2g |  | LEU159, TYR161, THR222 | −11.47 | 0.09 | N of Oxazole with H of NH of ALA162 (1.948) |
| 2h |  | VAL96, LEU159, THR222 | −11.24 | 0.11 | N of Oxazole with H of NH of ALA162 (1.687) |
| 2i |  | VAL96, LEU159, THR222 | −11.07 | 0.27 | N of Oxazole with H of NH of ALA162 (1.811) |
| 2k |  | VAL96, LEU159, THR222 | −10.84 | 0.26 | N of Oxazole with H of NH of ALA162 (2.004) |
| 2l |  | VAL96, LEU159, THR222 | −11.22 | 0.11 | N of Oxazole with H of NH of ALA162 (1.966) |
| 2m |  | VAL96, LEU159, THR222 | −10.88 | 0.13 | N of Oxazole with H of NH of ALA162 (1.967) |
| 2n |  | VAL96, LEU159, THR222 | −10.79 | 0.26 | N of Oxazole with H of NH of ALA162 (1.957) |
| Doxorubicin |  | VAL96, ALA162, THR222 | −12.92 | 0.89 | H of OH with O of Carbonyl of GLU166 (2.035) |
| Comp No. | Structures | Docked Amino Acid Residues (vdW Interactions) | Energy Score | RMSD Value | H Bond (Distance Å) |
|---|---|---|---|---|---|
| 1a |  | LEU1198, GLY1202, LEU1256 | −8.72 | 1.93 | O of Carbonyl with H of NH of MET1199 (1.932) |
| 1b |  | LEU1198, GLY1202, LEU1256 | −9.23 | 0.75 | None |
| 1c |  | LEU1198, GLY1202, LEU1256 | −9.17 | 0.84 | O of Carbonyl with H of NH of MET1199 (2.099) |
| 1d |  | MET1199, LEU1256, LEU1196, | −9.14 | 0.45 | H of NH with O of Carbonyl of GLU1197 (2.075) |
| 1e |  | LEU1196, LEU1198, LEU1256 | −8.65 | 1.91 | N of Thiazole with H of NH of MET1199 (2.221) |
| 1f |  | LEU1196, LEU1198, LEU1256 | −9.18 | 0.41 | N of Thiazole with H of NH of MET1199 (2.036) |
| 1g |  | ALA1148, LEU1198, ASP1203 | −8.99 | 1.87 | N of Thiazole with H of NH of MET1199 (2.158) |
| 1h |  | LEU1196, LEU1198, LEU1256 | −8.71 | 1.95 | N of Thiazole with H of NH of MET1199 (2.050) |
| 1i |  | LEU1122, GLU1197, LEU1256 | −9.65 | 0.28 | N of Thiazole with H of NH of MET1199 (2.041) |
| 1j |  | GLU1197, MET1199, ASP1203 | −8.70 | 0.17 | O of Carbonyl with H of NH of LYS1150 (2.056) |
| 1k |  | LEU1122, MET1199, LEU1256 | −8.47 | 0.77 | O of Carbonyl with H of NH of LYS1150 (1.986) |
| 1l |  | LEU1122, ALA1148, LEU1256 | −9.18 | 0.34 | H of NH with O of Carbonyl of MET1199 (2.122) |
| 1m |  | LEU1122, MET1199, LEU1256 | −8.49 | 1.84 | H of NH with O of Carbonyl of GLU1197 (2.143) |
| 1n |  | LEU1196, GLU1197, ASP1203 | −9.07 | 0.81 | H of NH with O of Carbonyl of MET1199 (1.895) |
| 1o |  | LEU1196, MET1199, LEU1256 | −9.38 | 0.50 | H of NH with O of Carbonyl of GLU1197 (2.117) |
| 2a |  | LEU1122, GLU1197, LEU1256 | −8.81 | 0.21 | N of Oxazole with H of NH of mMET1199 (1.810) |
| 2b |  | LEU1198, GLU1197, LEU1256 | −9.25 | 0.13 | N of Oxazole with H of NH of mMET1199 (1.742) |
| 2c |  | LEU1196, LEU1198, LEU1256 | −8.89 | 0.30 | N of Oxazole with H of NH of mMET1199 (1.758) |
| 2d |  | LEU1122, LEU2298, GLY1202 | −8.73 | 1.13 | N of Oxazole with H of NH of mMET1199 (1.758) |
| 2f |  | LEU1198, ASP1203, LEU1256 | −9.72 | 0.34 | N of Oxazole with H of NH of MET1199 (2.205) |
| 2g |  | LEU1198, ASP1203, LEU1256 | −9.75 | 1.34 | N of Oxazole with H of NH of MET1199 (2.968) |
| 2h |  | LEU1198, ASP1203, LEU1256 | −9.81 | 0.11 | N of Oxazole with H of NH of MET1199 (2.184) |
| 2i |  | LEU1198, ASP1203, LEU1256 | −9.42 | 0.77 | N of Oxazole with H of NH of MET1199 (1.840) |
| 2k |  | ALA1146, LEU1198, LEU1256 | 9.18 | 0.42 | N of Oxazole with H of NH of MET1199 (1.791) |
| 2l |  | LEU1198, GLU1197, LEU1256 | −8.74 | 1.54 | N of Oxazole with H of NH of MET1199 (1.973) |
| 2m |  | ALA1148, LEU1198, LEU1256 | −9.26 | 0.43 | N of Oxazole with H of NH of MET1199 (1.832) |
| 2n |  | ALA1148, LEU1198, LEU1256 | −9.18 | 0.12 | N of Oxazole with H of NH of MET1199 (1.780) |
| Crizotinib |  | LEU1196, LEU1198, MET1199 | −9.93 | 1.25 | None |
| Comp No. | Structures | Docked Amino Acid Residues (vdW Interactions) | Energy Score | RMSD Value | H Bond (Distance Å) |
|---|---|---|---|---|---|
| 1a |  | LEU855, LEU932, GLY993 | −9.68 | 1.11 | None |
| 1b |  | LEU855, LEU932, LEU983 | −10.08 | 1.10 | None |
| 1c |  | LEU855, LEU932, GLY993 | −9.68 | 1.38 | None |
| 1d |  | LEU855, LEU932, GLY993 | −9.76 | -0.29 | None |
| 1e |  | ALA880, LEU932, GLY993 | −9.47 | 0.87 | N of Thiazole with H of NH of LEU932 (2.066) |
| 1f |  | VAL863, ALA880, LEU932 | −9.65 | 0.49 | O of Carbonyl with H of NH of SER936 (2.089) |
| 1g |  | LEU855, LEU932, GLY993 | −9.39 | 1.73 | None |
| 1h |  | LEU855, LEU932, GLY993 | −9.87 | 0.39 | H of NH with O of Carbonyl of ASN961 (2.204) |
| 1i |  | LEU855, LEU932, LEU983 | −10.05 | 0.49 | None |
| 1j |  | LEU855, LEU932, LEU983 | −9.68 | 0.84 | None |
| 1k |  | LEU855, GLY935, LEU983 | −10.09 | 0.51 | O of Carbonyl with H of NH of LEU932 (1.919) |
| 1l |  | LEU855, GLY935, LEU983 | −10.57 | 0.93 | O of Carbonyl with H of NH of LEU932 (1.971) |
| 1m |  | GLY856, GLY861, GLY993 | −10.05 | 1.40 | H of NH with O of Carbonyl of ASN961 (2.215) |
| 1n |  | LEU855, TYR931, LEU983 | −10.02 | 0.49 | H of NH with O of Carbonyl of GLU930 (2.227) |
| 1o |  | LEU932, SER936, LEU983 | −9.19 | 0.19 | None |
| 2a |  | LEU855, MET929, LEU983 | −10.39 | 0.14 | N of Oxadiazole with H of NH of LEU932 (2.004) |
| 2b |  | LEU855, MET929, LEU983 | −10.70 | 0.11 | N of Oxadiazole with H of NH of LEU932 (2.042) |
| 2c |  | LEU855, MET929, LEU983 | −10.23 | 0.40 | N of Oxadiazole with H of NH of LEU932 (2.193) |
| 2d |  | LEU855, MET929, LEU983 | −10.22 | 0.11 | N of Oxadiazole with H of NH of LEU932 (2.051) |
| 2f |  | LEU932, SER936, LEU983 | −10.83 | 0.16 | None |
| 2g |  | LEU855, LEU932, LEU983 | −11.11 | 0.12 | N of Oxadiazole with H of NH of LEU932 (1.988) |
| 2h |  | LEU855, LEU932, LEU983 | −10.85 | 0.06 | N of Oxadiazole with H of NH of LEU932 (1.974) |
| 2i |  | LEU855, LEU932, LEU983 | −10.90 | 0.13 | N of Oxadiazole with H of NH of LEU932 (2.078) |
| 2k |  | LEU855, LEU932, LEU983 | −10.66 | 0.93 | N of Oxadiazole with H of NH of LEU932 (1.849) |
| 2l |  | LEU855, LEU932, LEU983 | −10.48 | 0.89 | None |
| 2m |  | LEU855, LEU932, LEU983 | −10.52 | 1.04 | N of Oxadiazole with H of NH of LEU932 (1.855) |
| 2n |  | LEU855, LEU932, LEU983 | −10.57 | 0.20 | N of Oxadiazole with H of NH of LEU932 (2.159) |
| Tamoxifen |  | LEU855, LEU983, GLY993 | −12.06 | 1.21 | None |
| Comp. No | MW (g/mol) a | LogP b | TPSA c | BBB d | GI Abs. e | Type of CYP Inh. f | Rule of Five g |
|---|---|---|---|---|---|---|---|
| 3a | 316.35 | 4.03 | 50.69 | Yes | High | CYP1A2, CYP2C19, CYP2C9, CYP2D6 | Yes |
| 3b | 350.80 | 4.58 | 50.69 | Yes | High | CYP1A2, CYP2C19, CYP2C9 | Yes |
| 3c | 317.34 | 3.13 | 63.58 | Yes | High | CYP1A2, CYP2C19, CYP2C9, CYP2D6 | Yes |
| 3d | 306.32 | 3.40 | 63.83 | Yes | High | CYP1A2, CYP2C19, CYP2C9, CYP2D6 | Yes |
| 3e | 338.38 | 3.19 | 104.71 | Yes | High | CYP1A2, CYP2C19, CYP2C9 | Yes |
| 3f | 341.36 | 3.87 | 74.48 | Yes | High | CYP1A2, CYP2C19, CYP2C9 | Yes |
| 3g | 375.81 | 4.42 | 74.48 | Yes | High | CYP1A2, CYP2C19, CYP2C9 | Yes |
| 3h | 342.35 | 2.90 | 87.37 | No | High | CYP1A2, CYP2C19, CYP2C9 | Yes |
| 3i | 331.32 | 3.17 | 87.62 | No | High | CYP1A2, CYP2C19, CYP2C9 | Yes |
| 3j | 363.39 | 2.98 | 97.27 | No | High | CYP1A2, CYP2C19, CYP2C9 | Yes |
| 3k | 350.80 | 4.64 | 50.69 | Yes | High | CYP1A2, CYP2C19, CYP2C9 | Yes |
| 3l | 385.24 | 5.17 | 50.69 | Yes | High | CYP1A2, CYP2C19, CYP2C9 | Yes |
| 3m | 351.79 | 3.65 | 63.58 | Yes | High | CYP1A2, CYP2C19, CYP2C9, CYP2D6, CYP3A4 | Yes |
| 3n | 340.76 | 3.83 | 63.83 | Yes | High | CYP1A2, CYP2C19, CYP2C9 | Yes |
| 3o | 372.83 | 3.71 | 104.71 | No | High | CYP1A2, CYP2C19, CYP2C9 | Yes |
| 4a | 314.34 | 4.36 | 48.15 | Yes | High | CYP1A2, CYP2C19, CYP2C9, CYP2D6 | Yes |
| 4b | 348.78 | 4.88 | 48.15 | No | High | CYP1A2, CYP2C19, CYP2C9, CYP2D6, | Yes |
| 4c | 315.33 | 3.58 | 61.04 | Yes | High | CYP1A2, CYP2C19, CYP2C9, CYP2D6, CYP3A4 | Yes |
| 4d | 304.30 | 3.66 | 61.29 | Yes | High | CYP1A2, CYP2C19, CYP2C9, CYP2D6, CYP3A4 | Yes |
| 4f | 339.35 | 4.04 | 71.94 | No | High | CYP1A2, CYP2C19, CYP2C9, CYP2D6, CYP3A4 | Yes |
| 4g | 373.79 | 4.62 | 71.94 | No | High | CYP1A2, CYP2C19, CYP2C9, CYP2D6, CYP3A4 | Yes |
| 4h | 340.33 | 3.39 | 84.83 | No | High | CYP1A2, CYP2C19, CYP2C9, CYP2D6, CYP3A4 | Yes |
| 4i | 329.31 | 3.45 | 85.08 | No | High | CYP1A2, CYP2C19, CYP2C9, CYP2D6, CYP3A4 | Yes |
| 4k | 348.78 | 4.73 | 48.15 | No | High | CYP1A2, CYP2C19, CYP2C9, CYP2D6, | Yes |
| 4l | 383.23 | 5.31 | 48.15 | No | High | CYP1A2, CYP2C19, CYP2C9 | Yes |
| 4m | 349.77 | 4.01 | 61.04 | Yes | High | CYP1A2, CYP2C19, CYP2C9, CYP2D6, CYP3A4 | Yes |
| 4n | 338.74 | 4.16 | 61.29 | No | High | CYP1A2, CYP2C19, CYP2C9, CYP2D6, CYP3A4 | Yes |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aydın, E.; Şentürk, A.M.; Küçük, H.B.; Güzel, M. Cytotoxic Activity and Docking Studies of 2-arenoxybenzaldehyde N-acyl Hydrazone and 1,3,4-Oxadiazole Derivatives against Various Cancer Cell Lines. Molecules 2022, 27, 7309. https://doi.org/10.3390/molecules27217309
Aydın E, Şentürk AM, Küçük HB, Güzel M. Cytotoxic Activity and Docking Studies of 2-arenoxybenzaldehyde N-acyl Hydrazone and 1,3,4-Oxadiazole Derivatives against Various Cancer Cell Lines. Molecules. 2022; 27(21):7309. https://doi.org/10.3390/molecules27217309
Chicago/Turabian StyleAydın, Esranur, Ahmet Mesut Şentürk, Hatice Başpınar Küçük, and Mustafa Güzel. 2022. "Cytotoxic Activity and Docking Studies of 2-arenoxybenzaldehyde N-acyl Hydrazone and 1,3,4-Oxadiazole Derivatives against Various Cancer Cell Lines" Molecules 27, no. 21: 7309. https://doi.org/10.3390/molecules27217309
APA StyleAydın, E., Şentürk, A. M., Küçük, H. B., & Güzel, M. (2022). Cytotoxic Activity and Docking Studies of 2-arenoxybenzaldehyde N-acyl Hydrazone and 1,3,4-Oxadiazole Derivatives against Various Cancer Cell Lines. Molecules, 27(21), 7309. https://doi.org/10.3390/molecules27217309