Discovery of Natural Small Molecules Promoting Collagen Secretion by High-Throughput Screening in Caenorhabditis elegans
Abstract
:1. Introduction
2. Results and Discussion
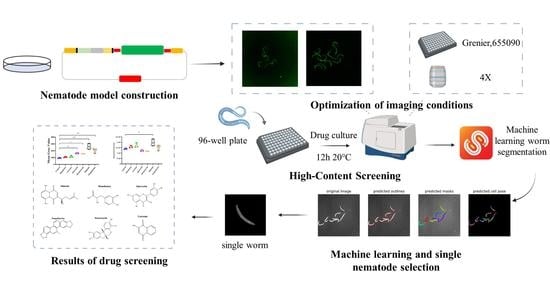
2.1. COL-12 Single Copy Insertion Transgenic Strain Construction in C. elegans
2.2. Development of High-Content Screening Approach for Prompting COL-12 Expression
2.3. Screening for Active Ingredients That Promote COL-12 Expression
2.4. Validation Study of the High Magnification Imaging Effect of Compounds Promoting COL-12 Expression
2.5. Validation of the Role of Natural Compounds
3. Materials and Methods
3.1. COL-12 Labelin g Worm Model Construction
3.2. Scellseg Software Application for Single Worm Cutting
3.3. Drug Screening and Data Analysis
3.4. Western Blotting
3.5. RT-PCR
3.6. Quantification and Statistical Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Gajbhiye, S.; Wairkar, S. Collagen fabricated delivery systems for wound healing: A new roadmap. Biomater. Adv. 2022, 142, 213152. [Google Scholar] [CrossRef] [PubMed]
- Gordon, M.K.; Hahn, R.A. Collagens. Cell Tissue Res. 2010, 339, 247–257. [Google Scholar] [CrossRef] [PubMed]
- Brodsky, B.; Persikov, A.V. Molecular structure of the collagen triple helix. Adv. Protein Chem. 2005, 70, 301–339. [Google Scholar] [PubMed]
- Chen, Q.; Wu, S.N.; Chen, Y.X.; Zhang, L.; Wei, H.Y.; Kumar, S.A. A novel missense COL10A1 mutation: C.2020G>A; p. Gly674Arg linked with the bowed legs stature in the Schmid metaphyseal chondrodysplasia-affected Chinese lineage. Bone Rep. 2019, 12, 100240. [Google Scholar] [CrossRef] [PubMed]
- Gouzoulis, M.J.; Kammien, A.J.; Zhu, J.R.; Gillinov, S.M.; Moore, H.G.; Grauer, J.N. Single-level posterior lumbar fusions in patients with Ehlers Danlos Syndrome not found to be associated with increased postoperative adverse events or five-year reoperations. N. Am. Spine Soc. J. 2022, 11, 100136. [Google Scholar] [PubMed]
- Marin, S.; Godet, I.; Nidadavolu, L.S.; Tian, J.; Dickinson, L.E.; Walston, J.D.; Gilkes, D.M.; Abadir, P.M. Valsartan and sacubitril combination treatment enhances collagen production in older adult human skin cells. Exp. Gerontol. 2022, 165, 111835. [Google Scholar] [CrossRef] [PubMed]
- Sabatelli, P.; Gualandi, F.; Gara, S.K.; Grumati, P.; Zamparelli, A.; Martoni, E.; Pellegrini, C.; Merlini, L.; Ferlini, A.; Bonaldo, P.; et al. Expression of collagen VI α5 and α6 chains in human muscle and in Duchenne muscular dystrophy-related muscle fibrosis. Matrix Biol. 2012, 31, 187–196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sabatelli, P.; Gara, S.K.; Grumati, P.; Urciuolo, A.; Gualandi, F.; Curci, R.; Squarzoni, S.; Zamparelli, A.; Martoni, E.; Merlini, L.; et al. Expression of the collagen VI α5 and α6 chains in normal human skin and in skin of patients with collagen VI-related myopathies. J. Investig. Dermatol. 2011, 131, 99–107. [Google Scholar] [CrossRef] [PubMed]
- Corsi, A.K.; Wightman, B.; Chalfie, M.A. Transparent Window into Biology: A Primer on Caenorhabditis elegans. Genetics 2015, 200, 387–407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brenner, S. The genetics of Caenorhabditis elegans. Genetics 1974, 77, 71–94. [Google Scholar] [CrossRef] [PubMed]
- Cox, G.N.; Kusch, M.; Edgar, R.S. Cuticle of Caenorhabditis elegans: Its isolation and partial characterization. J. Cell Biol. 1981, 90, 7–17. [Google Scholar] [CrossRef] [PubMed]
- Johnstone, I.L. Cuticle collagen genes. Expression in Caenorhabditis elegans. Trends Genet. 2000, 16, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Roberts, B.; Clucas, C.; Johnstone, I.L. Loss of SEC-23 in Caenorhabditis elegans causes defects in oogenesis, morphogenesis, and extracellular matrix secretion. Mol. Biol. Cell 2003, 14, 4414–4426. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shinn, P.; Chen, L.; Ferrer, M.; Itkin, Z.; Klumpp-Thomas, C.; McKnight, C.; Michael, S.; Mierzwa, T.; Thomas, C.; Wilson, K.; et al. High-Throughput Screening for Drug Combinations. Methods Mol. Biol. 2019, 1939, 11–35. [Google Scholar] [PubMed]
- Motoyaji, T. High-throughput Screening Technology for Selective Inhibitors of Transporters and Its Application in Drug Discovery. Yakugaku Zasshi J. Pharm. Soc. Jpn. 2021, 141, 511–515. [Google Scholar] [CrossRef] [PubMed]
- Frokjaer-Jensen, C.; Davis, M.W.; Hopkins, C.E.; Newman, B.J.; Thummel, J.M.; Olesen, S.P.; Grunnet, M.; Jorgensen, E.M. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat. Genet. 2008, 40, 1375–1383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xun, D.; Chen, D.; Zhou, Y.; Lauschke, Y.M.; Wang, R.; Wang, Y. Scellseg: A style-aware deep learning tool for adaptive cell instance segmentation by contrastive fine-tuning. IScience 2022, 25, 105506. [Google Scholar] [CrossRef] [PubMed]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fang, J.; Wu, X.; Meng, X.; Xun, D.; Xu, S.; Wang, Y. Discovery of Natural Small Molecules Promoting Collagen Secretion by High-Throughput Screening in Caenorhabditis elegans. Molecules 2022, 27, 8361. https://doi.org/10.3390/molecules27238361
Fang J, Wu X, Meng X, Xun D, Xu S, Wang Y. Discovery of Natural Small Molecules Promoting Collagen Secretion by High-Throughput Screening in Caenorhabditis elegans. Molecules. 2022; 27(23):8361. https://doi.org/10.3390/molecules27238361
Chicago/Turabian StyleFang, Jinyang, Xinyue Wu, Xi’nan Meng, Dejin Xun, Suhong Xu, and Yi Wang. 2022. "Discovery of Natural Small Molecules Promoting Collagen Secretion by High-Throughput Screening in Caenorhabditis elegans" Molecules 27, no. 23: 8361. https://doi.org/10.3390/molecules27238361
APA StyleFang, J., Wu, X., Meng, X., Xun, D., Xu, S., & Wang, Y. (2022). Discovery of Natural Small Molecules Promoting Collagen Secretion by High-Throughput Screening in Caenorhabditis elegans. Molecules, 27(23), 8361. https://doi.org/10.3390/molecules27238361