New Insight into the Substrate Selectivity of Bovine Milk γ-glutamyl Transferase via Structural and Molecular Dynamics Predictions
Abstract
:1. Introduction
2. Results and Discussion
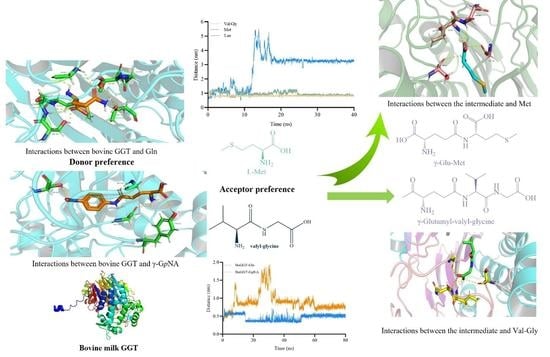
2.1. Mechanism behind the Donor Selectivity of BoGGT
2.1.1. Prediction of the 3D Structure of BoGGT
2.1.2. Interactions between BoGGT and Donor
2.1.3. MD Simulations of BoGGT–Donor Complex
2.2. Mechanism behind the Acceptor Selectivity of BoGGT
2.2.1. Construction and Optimization of the BoGGT Intermediate
2.2.2. Interactions between the BoGGT Intermediate and Acceptors
2.2.3. MD Simulations of BoGGT Intermediate–Acceptor Complex
3. Materials and Methods
3.1. Structure Construction of BoGGT and Molecular Docking between γ-glutamyl Donor and BoGGT
3.2. MD Simulation of BoGGT–Donor Complex
3.3. Construction of the BoGGT Intermediate and Optimization of the Intermediate
3.4. Molecular Docking between the BoGGT Intermediate and γ-glutamyl Acceptors
3.5. MD Simulation of BoGGT Intermediate–Acceptor Complexes
3.6. Statistical Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Cao, L.; Li, Q.; Lametsch, R. Identification and Activity Characterization of gamma-Glutamyltransferase from Bovine Milk. J. Agric. Food Chem. 2021, 69, 15325–15333. [Google Scholar] [CrossRef]
- Cao, L.; Li, Q.; Lametsch, R. Comparative analysis of substrate affinity and catalytic efficiency of gamma-glutamyltransferase from bovine milk and Bacillus amyloliquefaciens. Food Chem. 2023, 405 Pt B, 134930. [Google Scholar] [CrossRef]
- Hillmann, H.; Behr, J.; Ehrmann, M.A.; Vogel, R.F.; Hofmann, T. Formation of Kokumi-Enhancing gamma-Glutamyl Dipeptides in Parmesan Cheese by Means of gamma-Glutamyltransferase Activity and Stable Isotope Double-Labeling Studies. J. Agric. Food Chem. 2016, 64, 1784–1793. [Google Scholar] [CrossRef]
- Yang, J.; Bai, W.; Zeng, X.; Cui, C. Gamma glutamyl peptides: The food source, enzymatic synthesis, kokumi-active and the potential functional properties—A review. Trends Food Sci. Technol. 2019, 91, 339–346. [Google Scholar] [CrossRef]
- Zhao, C.J.; Schieber, A.; Ganzle, M.G. Formation of taste-active amino acids, amino acid derivatives and peptides in food fermentations—A review. Food Res. Int. 2016, 89 Pt 1, 39–47. [Google Scholar] [CrossRef]
- Lu, Y.; Wang, J.; Soladoye, O.P.; Aluko, R.E.; Fu, Y.; Zhang, Y. Preparation, receptors, bioactivity and bioavailability of γ-glutamyl peptides: A comprehensive review. Trends Food Sci. Technol. 2021, 113, 301–314. [Google Scholar] [CrossRef]
- Toelstede, S.; Hofmann, T. Kokumi-active glutamyl peptides in cheeses and their biogeneration by Penicillium roquefortii. J. Agric. Food Chem. 2009, 57, 3738–3748. [Google Scholar] [CrossRef] [PubMed]
- Toelstede, S.; Dunkel, A.; Hofmann, T. A series of kokumi peptides impart the long-lasting mouthfulness of matured Gouda cheese. J. Agric. Food Chem. 2009, 57, 1440–1448. [Google Scholar] [CrossRef]
- Yang, Z.; Wang, J.; Han, Z.; Blank, I.; Meng, F.; Wang, B.; Cao, Y.; Tian, H.; Chen, C. Isolation, identification and sensory evaluation of kokumi peptides from by-products of enzyme-modified butter. J. Sci. Food Agric. 2022, 102, 6668–6675. [Google Scholar] [CrossRef]
- Somma, V.; Calvio, C.; Rabuffetti, M.; Rama, E.; Speranza, G.; Morelli, C.F. An overall framework for the E. coli gamma-glutamyltransferase-catalyzed transpeptidation reactions. Bioorg. Chem. 2021, 115, 105217. [Google Scholar] [CrossRef]
- Shuai, Y.; Zhang, T.; Jiang, B.; Mu, W. Development of efficient enzymatic production of theanine by gamma-glutamyltranspeptidase from a newly isolated strain of Bacillus subtilis, SK11.004. J. Sci. Food Agric. 2010, 90, 2563–2567. [Google Scholar] [CrossRef]
- Blel, M.; Guingamp, M.-F.; Gaillard, J.-L.; Humbert, G. Studies on the thermal sensitivity of γ-glutamyl transpeptidase measured with a modified test procedure and compared with that of alkaline phosphatase and lactoperoxidase in milk. Le Lait 2002, 82, 555–566. [Google Scholar] [CrossRef] [Green Version]
- Baumrucker, C.R. gamma-Glutamyl transpeptidase of bovine milk membranes: Distribution and characterization. J. Dairy Sci. 1979, 62, 253–258. [Google Scholar] [CrossRef] [PubMed]
- Tate, S.S.; Meister, A. Interaction of gamma-glutamyl transpeptidase with amino acids, dipeptides, and derivatives and analogs of glutathione. J. Biol. Chem. 1974, 249, 7593–7602. [Google Scholar] [CrossRef]
- West, M.B.; Chen, Y.; Wickham, S.; Heroux, A.; Cahill, K.; Hanigan, M.H.; Mooers, B.H. Novel insights into eukaryotic gamma-glutamyltranspeptidase 1 from the crystal structure of the glutamate-bound human enzyme. J. Biol. Chem. 2013, 288, 31902–31913. [Google Scholar] [CrossRef] [Green Version]
- Terzyan, S.S.; Burgett, A.W.; Heroux, A.; Smith, C.A.; Mooers, B.H.; Hanigan, M.H. Human gamma-Glutamyl Transpeptidase 1: Structures of the free enzyme, inhibitor-bound tetrahedral transition states, and glutamate-bound enzyme reveal novel movement within the active site during catalysis. J. Biol. Chem. 2015, 290, 17576–17586. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taylor, R.D.; Jewsbury, P.J.; Essex, J.W. A review of protein-small molecule docking methods. J. Comput. Aided Mol. Des. 2002, 16, 151–166. [Google Scholar] [CrossRef] [PubMed]
- Sledz, P.; Caflisch, A. Protein structure-based drug design: From docking to molecular dynamics. Curr. Opin. Struct. Biol. 2018, 48, 93–102. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Zidek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Castonguay, R.; Halim, D.; Morin, M.; Furtos, A.; Lherbet, C.; Bonneil, E.; Thibault, P.; Keillor, J.W. Kinetic characterization and identification of the acylation and glycosylation sites of recombinant human gamma-glutamyltranspeptidase. Biochemistry 2007, 46, 12253–12262. [Google Scholar] [CrossRef]
- Ikeda, Y.; Fujii, J.; Anderson, M.E.; Taniguchi, N.; Meister, A. Involvement of Ser-451 and Ser-452 in the catalysis of human gamma-glutamyl transpeptidase. J. Biol. Chem. 1995, 270, 22223–22228. [Google Scholar] [CrossRef] [Green Version]
- Chen, D.; Oezguen, N.; Urvil, P.; Ferguson, C.; Dann, S.M.; Savidge, T.C. Regulation of protein-ligand binding affinity by hydrogen bond pairing. Sci. Adv. 2016, 2, e1501240. [Google Scholar] [CrossRef] [Green Version]
- Hibi, T.; Nii, H.; Nakatsu, T.; Kimura, A.; Kato, H.; Hiratake, J.; Oda, J. Crystal structure of gamma-glutamylcysteine synthetase: Insights into the mechanism of catalysis by a key enzyme for glutathione homeostasis. Proc. Natl. Acad. Sci. USA 2004, 101, 15052–15057. [Google Scholar] [CrossRef] [Green Version]
- Okada, T.; Suzuki, H.; Wada, K.; Kumagai, H.; Fukuyama, K. Crystal structures of gamma-glutamyltranspeptidase from Escherichia coli, a key enzyme in glutathione metabolism, and its reaction intermediate. Proc. Natl. Acad. Sci. USA 2006, 103, 6471–6476. [Google Scholar] [CrossRef] [Green Version]
- Morrow, A.L.; Williams, K.; Sand, A.; Boanca, G.; Barycki, J.J. Characterization of Helicobacter pylori gamma-glutamyltranspeptidase reveals the molecular basis for substrate specificity and a critical role for the tyrosine 433-containing loop in catalysis. Biochemistry 2007, 46, 13407–13414. [Google Scholar] [CrossRef]
- Wada, K.; Irie, M.; Suzuki, H.; Fukuyama, K. Crystal structure of the halotolerant gamma-glutamyltranspeptidase from Bacillus subtilis in complex with glutamate reveals a unique architecture of the solvent-exposed catalytic pocket. FEBS J. 2010, 277, 1000–1009. [Google Scholar] [CrossRef]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [Green Version]
- Eberhardt, J.; Santos-Martins, D.; Tillack, A.F.; Forli, S. AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings. J. Chem. Inf. Model. 2021, 61, 3891–3898. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Hunt, C.J.; Lin, S.; Meyer, A.S.; Li, Q.; Lametsch, R. Elucidation of the Molecular Mechanism of Bovine Milk gamma-Glutamyltransferase Catalyzed Formation of gamma-Glutamyl-Valyl-Glycine. J. Agric. Food Chem. 2023, 71, 2455–2463. [Google Scholar] [CrossRef] [PubMed]
- Abdulrashid, N.I.; Aminu, S.; Adamu, R.M.; Tajuddeen, N.; Isah, M.B.; Jatau, I.D.; Aliyu, A.B.; Simelane, M.B.C.; Onyike, E.; Ibrahim, M.A. Phloroglucinol as a Potential Candidate against Trypanosoma congolense Infection: Insights from In Vivo, In Vitro, Molecular Docking and Molecular Dynamic Simulation Analyses. Molecules 2022, 27, 469. [Google Scholar] [CrossRef] [PubMed]









| Complex | Total Binding Energy (kJ/mol) | Van der Waals Energy (kJ/mol) | Electrostatic Energy (kJ/mol) | Polar Solvation Energy (kJ/mol) | SASA Energy (kJ/mol) |
|---|---|---|---|---|---|
| BoGGT intermediate–Val-Gly | −141.062 ± 14.829 | −176.361 ± 25.742 | −102.037 ± 36.825 | 165.193 ± 41.980 | −27.857 ± 5.184 |
| BoGGT intermediate–Met | −110.784 ± 11.275 | −157.906 ± 32.609 | −76.074 ± 23.259 | 134.209 ± 33.527 | −11.013 ± 4.513 |
| BoGGT intermediate–Leu | −75.481 ± 19.973 | −123.524 ± 27.128 | −62.598 ± 19.937 | 124.372 ± 32.252 | −13.731 ± 4.235 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cao, L.; Hunt, C.J.; Meyer, A.S.; Lametsch, R. New Insight into the Substrate Selectivity of Bovine Milk γ-glutamyl Transferase via Structural and Molecular Dynamics Predictions. Molecules 2023, 28, 4657. https://doi.org/10.3390/molecules28124657
Cao L, Hunt CJ, Meyer AS, Lametsch R. New Insight into the Substrate Selectivity of Bovine Milk γ-glutamyl Transferase via Structural and Molecular Dynamics Predictions. Molecules. 2023; 28(12):4657. https://doi.org/10.3390/molecules28124657
Chicago/Turabian StyleCao, Lichuang, Cameron J. Hunt, Anne S. Meyer, and René Lametsch. 2023. "New Insight into the Substrate Selectivity of Bovine Milk γ-glutamyl Transferase via Structural and Molecular Dynamics Predictions" Molecules 28, no. 12: 4657. https://doi.org/10.3390/molecules28124657
APA StyleCao, L., Hunt, C. J., Meyer, A. S., & Lametsch, R. (2023). New Insight into the Substrate Selectivity of Bovine Milk γ-glutamyl Transferase via Structural and Molecular Dynamics Predictions. Molecules, 28(12), 4657. https://doi.org/10.3390/molecules28124657