Protein-Based Hydrogels and Their Biomedical Applications
Abstract
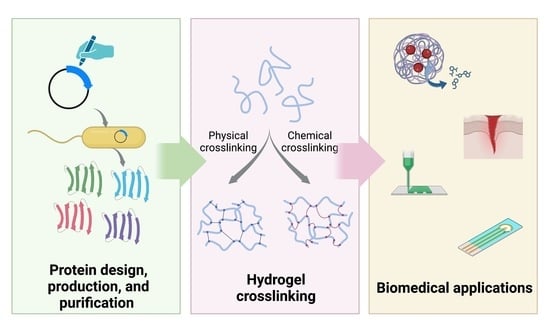
:1. Introduction
2. Hydrogels Crosslinking Strategies
2.1. Physically Crosslinked Protein Hydrogel
2.1.1. pH and Ion-Induced Protein Hydrogels
2.1.2. Temperature-Induced Hydrogel Formation
2.1.3. Protein–Protein Interaction-Induced Assembly of Hydrogels
2.2. Chemically Crosslinked Protein Hydrogel
2.2.1. Light-Controlled Protein Hydrogel Formation
2.2.2. Chemical Crosslinker-Based Protein Hydrogels
2.2.3. Enzymatic Crosslinked Hydrogels
3. Hydrogels Made from Different Protein Sources and Their Biomedical Applications
3.1. Hydrogels Made of Natural Proteins
3.2. Hydrogels Made of Microbially Synthesized Proteins
3.2.1. Strategies for Microbial Synthesis of Material Proteins
3.2.2. Microbially Synthesize Proteins of Different Origins and Their Applications
3.2.3. Multi-Functional Hydrogels from Different Sources
| Application | Protein | Desirable Properties | References |
|---|---|---|---|
| Bioadhesion | Mussel-foot protein |
| [62] |
| Drug delivery | Elastin-like polypeptideKeratin |
| [97,98] |
| Wound healing | Suckerin–spider silkKeratin |
| [92,99] |
| 3D cell culture | Spider silkElastin–collagenKeratin |
| [91,100,101] |
| Tissue regeneration | CollagenKeratin–fibrinogen |
| [77,83,84] |
| Biofabrication | Spider silkCollagen |
| [80,91] |
| Wearable sensor | Resilin |
| [93] |
4. Current Challenges and Future Directions
4.1. Synthetic Biology for Material Production and Protein Hydrogel Innovations
4.2. Sequence–Structure–Function Relationships
4.3. Lack of Anisotropy in Protein Hydrogels
4.4. Dynamic Cell–Hydrogel Interactions
4.5. Computation-Assisted Approaches
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Wichterle, O.; Lím, D. Hydrophilic Gels for Biological Use. Nature 1960, 185, 117–118. [Google Scholar] [CrossRef]
- Mantha, S.; Pillai, S.; Khayambashi, P.; Upadhyay, A.; Zhang, Y.; Tao, O.; Pham, H.M.; Tran, S.D. Smart Hydrogels in Tissue Engineering and Regenerative Medicine. Materials 2019, 12, 3323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vigata, M.; Meinert, C.; Hutmacher, D.W.; Bock, N. Hydrogels as Drug Delivery Systems: A Review of Current Characterization and Evaluation Techniques. Pharmaceutics 2020, 12, 1188. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.; Jeon, J.; Zhu, Y.; Hoppe, E.D.; Jun, Y.-S.; Genin, G.M.; Zhang, F. A Biosynthetic Hybrid Spidroin-Amyloid-Mussel Foot Protein for Underwater Adhesion on Diverse Surfaces. ACS Appl. Mater. Interfaces 2021, 13, 48457–48468. [Google Scholar] [CrossRef] [PubMed]
- Haiyan, Z.; Baoqi, Z. Functional Silk Fibroin Hydrogels: Preparation, Properties and Applications. J. Mater. Chem. B 2021, 9, 1238. [Google Scholar] [CrossRef]
- Antoine, E.E.; Vlachos, P.P.; Rylander, M.N. Review of Collagen I Hydrogels for Bioengineered Tissue Microenvironments: Characterization of Mechanics, Structure, and Transport. Tissue Eng. Part B Rev. 2014, 20, 683–696. [Google Scholar] [CrossRef] [Green Version]
- Sarrigiannidis, S.O.; Rey, J.M.; Dobre, O.; González-García, C.; Dalby, M.J.; Salmeron-Sanchez, M. A Tough Act to Follow: Collagen Hydrogel Modifications to Improve Mechanical and Growth Factor Loading Capabilities. Mater. Today Bio 2021, 10, 100098. [Google Scholar] [CrossRef]
- Gibson, D.G.; Young, L.; Chuang, R.-Y.; Venter, J.C.; Hutchison, C.A.; Smith, H.O. Enzymatic Assembly of DNA Molecules up to Several Hundred Kilobases. Nat. Methods 2009, 6, 343–345. [Google Scholar] [CrossRef]
- Engler, C.; Kandzia, R.; Marillonnet, S. A One Pot, One Step, Precision Cloning Method with High Throughput Capability. PLoS ONE 2008, 3, e3647. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Kong, N.; Laver, B.; Liu, J. Hydrogels Constructed from Engineered Proteins. Small 2016, 12, 973–987. [Google Scholar] [CrossRef]
- Zhu, Z.; Ling, S.; Yeo, J.; Zhao, S.; Tozzi, L.; Buehler, M.J.; Omenetto, F.; Li, C.; Kaplan, D.L. High-Strength, Durable All-Silk Fibroin Hydrogels with Versatile Processability toward Multifunctional Applications. Adv. Funct. Mater. 2018, 28, 1704757. [Google Scholar] [CrossRef]
- Ahn, W.; Lee, J.-H.; Kim, S.R.; Lee, J.; Lee, E.J. Designed Protein- and Peptide-Based Hydrogels for Biomedical Sciences. J. Mater. Chem. B 2021, 9, 1919–1940. [Google Scholar] [CrossRef]
- Hennink, W.E.; van Nostrum, C.F. Novel Crosslinking Methods to Design Hydrogels. Adv. Drug Deliv. Rev. 2012, 64, 223–236. [Google Scholar] [CrossRef]
- Wang, H.; Shi, Y.; Wang, L.; Yang, Z. Recombinant Proteins as Cross-Linkers for Hydrogelations. Chem. Soc. Rev. 2013, 42, 891–901. [Google Scholar] [CrossRef]
- Nie, J.; Zhang, X.; Wang, W.; Ren, J.; Zeng, A.-P. Tunable Protein Hydrogels: Present State and Emerging Development. In Tunable Hydrogels; Lavrentieva, A., Pepelanova, I., Seliktar, D., Eds.; Advances in Biochemical Engineering/Biotechnology; Springer International Publishing: Cham, Switzerland, 2021; Volume 178, pp. 63–97. ISBN 978-3-030-76768-6. [Google Scholar]
- Hollingshead, S.; Lin, C.; Liu, J.C. Designing Smart Materials with Recombinant Proteins. Macromol. Biosci. 2017, 17, 1600554. [Google Scholar] [CrossRef]
- Mie, M.; Oomuro, M.; Kobatake, E. Hydrogel Scaffolds Composed of Genetically Synthesized Self-Assembling Peptides for Three-Dimensional Cell Culture. Polym. J. 2013, 45, 504–508. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Zhang, F. Amyloids as Building Blocks for Macroscopic Functional Materials: Designs, Applications and Challenges. Int. J. Mol. Sci. 2021, 22, 10698. [Google Scholar] [CrossRef]
- Narayanaswamy, R.; Torchilin, V.P. Hydrogels and Their Applications in Targeted Drug Delivery. Molecules 2019, 24, 603. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mithieux, S.M.; Wise, S.G.; Weiss, A.S. Tropoelastin—A Multifaceted Naturally Smart Material. Adv. Drug Deliv. Rev. 2013, 65, 421–428. [Google Scholar] [CrossRef]
- Wise, S.G.; Yeo, G.C.; Hiob, M.A.; Rnjak-Kovacina, J.; Kaplan, D.L.; Ng, M.K.C.; Weiss, A.S. Tropoelastin—A Versatile, Bioactive Assembly Module. Acta Biomater. 2014, 10, 1532–1541. [Google Scholar] [CrossRef] [Green Version]
- Mithieux, S.M.; Tu, Y.; Korkmaz, E.; Braet, F.; Weiss, A.S. In Situ Polymerization of Tropoelastin in the Absence of Chemical Cross-Linking. Biomaterials 2009, 30, 431–435. [Google Scholar] [CrossRef] [PubMed]
- Luo, F.; Qian, Z.-G.; Xia, X.-X. Responsive Protein Hydrogels Assembled from Spider Silk Carboxyl-Terminal Domain and Resilin Copolymers. Polymers 2018, 10, 915. [Google Scholar] [CrossRef] [Green Version]
- Garcia Garcia, C.; Patkar, S.S.; Wang, B.; Abouomar, R.; Kiick, K.L. Recombinant Protein-Based Injectable Materials for Biomedical Applications. Adv. Drug Deliv. Rev. 2023, 193, 114673. [Google Scholar] [CrossRef]
- Parker, R.N.; Cairns, D.M.; Wu, W.A.; Jordan, K.; Guo, C.; Huang, W.; Martin-Moldes, Z.; Kaplan, D.L. Smart Material Hydrogel Transfer Devices Fabricated with Stimuli-Responsive Silk-Elastin-Like Proteins. Adv. Healthcare Mater. 2020, 9, 2000266. [Google Scholar] [CrossRef]
- Shi, H.; Ji, T.; Zhai, C.; Lu, J.; Huang, W.; Yeo, J. Thermo- and Ion-Responsive Silk-Elastin-like Proteins and Their Multiscale Mechanisms. J. Mater. Chem. B 2022, 10, 6133–6142. [Google Scholar] [CrossRef]
- Balu, R.; Dutta, N.K.; Dutta, A.K.; Choudhury, N.R. Resilin-Mimetics as a Smart Biomaterial Platform for Biomedical Applications. Nat. Commun. 2021, 12, 149. [Google Scholar] [CrossRef]
- Dutta, N.K.; Truong, M.Y.; Mayavan, S.; Roy Choudhury, N.; Elvin, C.M.; Kim, M.; Knott, R.; Nairn, K.M.; Hill, A.J. A Genetically Engineered Protein Responsive to Multiple Stimuli. Angew. Chem. Int. Ed. 2011, 50, 4428–4431. [Google Scholar] [CrossRef] [PubMed]
- Andrade, F.; Roca-Melendres, M.M.; Durán-Lara, E.F.; Rafael, D.; Schwartz, S. Stimuli-Responsive Hydrogels for Cancer Treatment: The Role of PH, Light, Ionic Strength and Magnetic Field. Cancers 2021, 13, 1164. [Google Scholar] [CrossRef]
- Macias, M.J.; Oschkinat, H.; Gervais, V.; Civera, C. Structural Analysis of WW Domains and Design of a WW Prototype. Nat. Struct. Biol. 2000, 7, 375–379. [Google Scholar] [CrossRef] [PubMed]
- Wong Po Foo, C.T.S.; Lee, J.S.; Mulyasasmita, W.; Parisi-Amon, A.; Heilshorn, S.C. Two-Component Protein-Engineered Physical Hydrogels for Cell Encapsulation. Proc. Natl. Acad. Sci. USA 2009, 106, 22067–22072. [Google Scholar] [CrossRef] [Green Version]
- Hu, W.; Wang, Z.; Xiao, Y.; Zhang, S.; Wang, J. Advances in Crosslinking Strategies of Biomedical Hydrogels. Biomater. Sci. 2019, 7, 843–855. [Google Scholar] [CrossRef]
- Narayan, O.P.; Mu, X.; Hasturk, O.; Kaplan, D.L. Dynamically Tunable Light Responsive Silk-Elastin-like Proteins. Acta Biomater. 2021, 121, 214–223. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Su, C.; Chen, Y.; Tian, S.; Lu, C.; Huang, W.; Lv, Q. Current Understanding of the Applications of Photocrosslinked Hydrogels in Biomedical Engineering. Gels 2022, 8, 216. [Google Scholar] [CrossRef]
- Li, L.; Scheiger, J.M.; Levkin, P.A. Design and Applications of Photoresponsive Hydrogels. Adv. Mater. 2019, 31, 1807333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Su, J.; Li, J.; Liang, J.; Zhang, K.; Li, J. Hydrogel Preparation Methods and Biomaterials for Wound Dressing. Life 2021, 11, 1016. [Google Scholar] [CrossRef]
- Eke, G.; Mangir, N.; Hasirci, N.; MacNeil, S.; Hasirci, V. Development of a UV Crosslinked Biodegradable Hydrogel Containing Adipose Derived Stem Cells to Promote Vascularization for Skin Wounds and Tissue Engineering. Biomaterials 2017, 129, 188–198. [Google Scholar] [CrossRef]
- Kim, S.H.; Yeon, Y.K.; Lee, J.M.; Chao, J.R.; Lee, Y.J.; Seo, Y.B.; Sultan, M.T.; Lee, O.J.; Lee, J.S.; Yoon, S.; et al. Precisely Printable and Biocompatible Silk Fibroin Bioink for Digital Light Processing 3D Printing. Nat. Commun. 2018, 9, 1620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, Z.; Xu, Y.; Dong, L.; Desai, M.S.; Xia, J.; Liang, M.; Lee, S.-W.; Mi, S.; Sun, W. Design of Functional Hydrogels Using Smart Polymer Based on Elastin-like Polypeptides. Chem. Eng. J. 2022, 435, 135155. [Google Scholar] [CrossRef]
- Hadar, D.; Strugach, D.S.; Amiram, M. Conjugates of Recombinant Protein-Based Polymers: Combining Precision with Chemical Diversity. Adv. NanoBiomed Res. 2022, 2, 2100142. [Google Scholar] [CrossRef]
- Madl, C.M.; Heilshorn, S.C. Rapid Diels–Alder Cross-Linking of Cell Encapsulating Hydrogels. Chem. Mater. 2019, 31, 8035–8043. [Google Scholar] [CrossRef]
- Pal, K.; Paulson, A.T.; Rousseau, D. 14—Biopolymers in Controlled-Release Delivery Systems. In Handbook of Biopolymers and Biodegradable Plastics; Ebnesajjad, S., Ed.; Plastics Design Library; William Andrew Publishing: Boston, MA, USA, 2013; pp. 329–363. ISBN 978-1-4557-2834-3. [Google Scholar]
- Cengiz, N.; Gevrek, T.N.; Sanyal, R.; Sanyal, A. Fabrication of Patterned Hydrogel Interfaces: Exploiting the Maleimide Group as a Dual Purpose Handle for Cross-Linking and Bioconjugation. Bioconjugate Chem. 2020, 31, 1382–1391. [Google Scholar] [CrossRef] [PubMed]
- Hua, J.; Li, Z.; Xia, W.; Yang, N.; Gong, J.; Zhang, J.; Qiao, C. Preparation and Properties of EDC/NHS Mediated Crosslinking Poly (Gamma-Glutamic Acid)/Epsilon-Polylysine Hydrogels. Mater. Sci. Eng. C 2016, 61, 879–892. [Google Scholar] [CrossRef]
- Chimisso, V.; Aleman Garcia, M.A.; Yorulmaz Avsar, S.; Dinu, I.A.; Palivan, C.G. Design of Bio-Conjugated Hydrogels for Regenerative Medicine Applications: From Polymer Scaffold to Biomolecule Choice. Molecules 2020, 25, 4090. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Tian, J.; Mao, H.; Gu, Z. Bioadhesives: Current Hotspots and Emerging Challenges. Curr. Opin. Biomed. Eng. 2021, 18, 100271. [Google Scholar] [CrossRef]
- Jeon, J.; Subramani, S.V.; Lee, K.Z.; Jiang, B.; Zhang, F. Microbial Synthesis of High-Molecular-Weight, Highly Repetitive Protein Polymers. Int. J. Mol. Sci. 2023, 24, 6416. [Google Scholar] [CrossRef]
- Li, Y.; Xue, B.; Cao, Y. 100th Anniversary of Macromolecular Science Viewpoint: Synthetic Protein Hydrogels. ACS Macro Lett. 2020, 9, 512–524. [Google Scholar] [CrossRef]
- McHale, M.K.; Setton, L.A.; Chilkoti, A. Synthesis and in Vitro Evaluation of Enzymatically Cross-Linked Elastin-Like Polypeptide Gels for Cartilaginous Tissue Repair. Tissue Eng. 2005, 11, 1768–1779. [Google Scholar] [CrossRef]
- Li, Y.; Su, J.; Cavaco-Paulo, A. Laccase-Catalyzed Cross-Linking of BSA Mediated by Tyrosine. Int. J. Biol. Macromol. 2021, 166, 798–805. [Google Scholar] [CrossRef]
- Wang, S.; Huang, W.; Feng, Z.; Tian, X.; Wang, D.; Rao, L.; Tan, M.; Roongsawang, N.; Song, H.; Jiang, W.; et al. Laccase-Mediated Formation of Hydrogels Based on Silk-Elastin-like Protein Polymers with Ultra-High Molecular Weight. Int. J. Biol. Macromol. 2023, 231, 123239. [Google Scholar] [CrossRef]
- Kou, S.; Yang, Z.; Luo, J.; Sun, F. Entirely Recombinant Protein-Based Hydrogels for Selective Heavy Metal Sequestration. Polym. Chem. 2017, 8, 6158–6164. [Google Scholar] [CrossRef]
- Sood, A.; Ji, S.M.; Kumar, A.; Han, S.S. Enzyme-Triggered Crosslinked Hybrid Hydrogels for Bone Tissue Engineering. Materials 2022, 15, 6383. [Google Scholar] [CrossRef]
- Schneider, A.; Garlick, J.A.; Egles, C. Self-Assembling Peptide Nanofiber Scaffolds Accelerate Wound Healing. PLoS ONE 2008, 3, e1410. [Google Scholar] [CrossRef] [Green Version]
- Gelain, F.; Horii, A.; Zhang, S. Designer Self-Assembling Peptide Scaffolds for 3-D Tissue Cell Cultures and Regenerative Medicine. Macromol. Biosci. 2007, 7, 544–551. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, B.J.D.; Couto, R.D. Toxicological Alert: Exposure to Glycidyl Methacrylate and Cancer Risk. Toxicol. Ind. Health 2020, 36, 937–939. [Google Scholar] [CrossRef]
- Kam, D.; Olender, A.; Rudich, A.; Kan-Tor, Y.; Buxboim, A.; Shoseyov, O.; Magdassi, S. 3D Printing of Resilin in Water by Multiphoton Absorption Polymerization. Adv. Funct. Mater. 2023, 2210993. [Google Scholar] [CrossRef]
- Charati, M.B.; Ifkovits, J.L.; Burdick, J.A.; Linhardt, J.G.; Kiick, K.L. Hydrophilic Elastomeric Biomaterials Based on Resilin-like Polypeptides. Soft Matter 2009, 5, 3412–3416. [Google Scholar] [CrossRef] [Green Version]
- Lim, D.W.; Nettles, D.L.; Setton, L.A.; Chilkoti, A. Rapid Crosslinking of Elastin-like Polypeptides with Hydroxymethylphosphines in Aqueous Solution. Biomacromolecules 2007, 8, 1463–1470. [Google Scholar] [CrossRef] [Green Version]
- Dai, B.; Sargent, C.J.; Gui, X.; Liu, C.; Zhang, F. Fibril Self-Assembly of Amyloid–Spider Silk Block Polypeptides. Biomacromolecules 2019, 20, 2015–2023. [Google Scholar] [CrossRef]
- Li, J.; Zhu, Y.; Yu, H.; Dai, B.; Jun, Y.-S.; Zhang, F. Microbially Synthesized Polymeric Amyloid Fiber Promotes β-Nanocrystal Formation and Displays Gigapascal Tensile Strength. ACS Nano 2021, 15, 11843–11853. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.; Dai, B.; Qiao, J.B.; Li, W.; Fortner, J.D.; Zhang, F. Microbially Synthesized Repeats of Mussel Foot Protein Display Enhanced Underwater Adhesion. ACS Appl. Mater. Interfaces 2018, 10, 43003–43012. [Google Scholar] [CrossRef]
- Bai, W.; Sargent, C.J.; Choi, J.-M.; Pappu, R.V.; Zhang, F. Covalently-Assembled Single-Chain Protein Nanostructures with Ultra-High Stability. Nat. Commun. 2019, 10, 3317. [Google Scholar] [CrossRef] [Green Version]
- Bowen, C.H.; Reed, T.J.; Sargent, C.J.; Mpamo, B.; Galazka, J.M.; Zhang, F. Seeded Chain-Growth Polymerization of Proteins in Living Bacterial Cells. ACS Synth. Biol. 2019, 8, 2651–2658. [Google Scholar] [CrossRef] [PubMed]
- Bowen, C.H.; Sargent, C.J.; Wang, A.; Zhu, Y.; Chang, X.; Li, J.; Mu, X.; Galazka, J.M.; Jun, Y.-S.; Keten, S.; et al. Microbial Production of Megadalton Titin Yields Fibers with Advantageous Mechanical Properties. Nat. Commun. 2021, 12, 5182. [Google Scholar] [CrossRef] [PubMed]
- Bowen, C.H.; Dai, B.; Sargent, C.J.; Bai, W.; Ladiwala, P.; Feng, H.; Huang, W.; Kaplan, D.L.; Galazka, J.M.; Zhang, F. Recombinant Spidroins Fully Replicate Primary Mechanical Properties of Natural Spider Silk. Biomacromolecules 2018, 19, 3853–3860. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmuck, B.; Greco, G.; Barth, A.; Pugno, N.M.; Johansson, J.; Rising, A. High-Yield Production of a Super-Soluble Miniature Spidroin for Biomimetic High-Performance Materials. Mater. Today 2021, 50, 16–23. [Google Scholar] [CrossRef]
- Li, J.; Jiang, B.; Chang, X.; Yu, H.; Han, Y.; Zhang, F. Bi-Terminal Fusion of Intrinsically-Disordered Mussel Foot Protein Fragments Boosts Mechanical Strength for Protein Fibers. Nat. Commun. 2023, 14, 2127. [Google Scholar] [CrossRef]
- McCormick, R.J. The Flexibility of the Collagen Compartment of Muscle. Meat Sci. 1994, 36, 79–91. [Google Scholar] [CrossRef]
- Buehler, M.J. Atomistic and Continuum Modeling of Mechanical Properties of Collagen: Elasticity, Fracture, and Self-Assembly. J. Mater. Res. 2006, 21, 1947–1961. [Google Scholar] [CrossRef] [Green Version]
- Shen, Z.L.; Kahn, H.; Ballarini, R.; Eppell, S.J. Viscoelastic Properties of Isolated Collagen Fibrils. Biophys. J. 2011, 100, 3008–3015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Potekhin, S.A.; Senin, A.A.; Abdurakhmanov, N.N.; Tiktopulo, E.I. High Pressure Stabilization of Collagen Structure. Biochim. Et Biophys. Acta (BBA) Proteins Proteom. 2009, 1794, 1151–1158. [Google Scholar] [CrossRef]
- Myllyharju, J.; Nokelainen, M.; Vuorela, A.; Kivirikko, K.I. Expression of Recombinant Human Type I-III Collagens in the Yeast Pichia Pastoris. Biochem. Soc. Trans. 2000, 28, 353–357. [Google Scholar] [CrossRef] [PubMed]
- Olsen, D.; Yang, C.; Bodo, M.; Chang, R.; Leigh, S.; Baez, J.; Carmichael, D.; Perälä, M.; Hämäläinen, E.-R.; Jarvinen, M.; et al. Recombinant Collagen and Gelatin for Drug Delivery. Adv. Drug Deliv. Rev. 2003, 55, 1547–1567. [Google Scholar] [CrossRef]
- Elsdale, T.; Bard, J. Collagen Substrata for Studies on Cell Behavior. J. Cell Biol. 1972, 54, 626–637. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, C.; Hillas, P.J.; Báez, J.A.; Nokelainen, M.; Balan, J.; Tang, J.; Spiro, R.; Polarek, J.W. The Application of Recombinant Human Collagen in Tissue Engineering. BioDrugs 2004, 18, 103–119. [Google Scholar] [CrossRef] [PubMed]
- Ramírez-Rodríguez, G.B.; Montesi, M.; Panseri, S.; Sprio, S.; Tampieri, A.; Sandri, M. Biomineralized Recombinant Collagen-Based Scaffold Mimicking Native Bone Enhances Mesenchymal Stem Cell Interaction and Differentiation. Tissue Eng. Part A 2017, 23, 1423–1435. [Google Scholar] [CrossRef] [PubMed]
- Tytgat, L.; Dobos, A.; Markovic, M.; Van Damme, L.; Van Hoorick, J.; Bray, F.; Thienpont, H.; Ottevaere, H.; Dubruel, P.; Ovsianikov, A.; et al. High-Resolution 3D Bioprinting of Photo-Cross-Linkable Recombinant Collagen to Serve Tissue Engineering Applications. Biomacromolecules 2020, 21, 3997–4007. [Google Scholar] [CrossRef]
- Liu, W.; Merrett, K.; Griffith, M.; Fagerholm, P.; Dravida, S.; Heyne, B.; Scaiano, J.C.; Watsky, M.A.; Shinozaki, N.; Lagali, N.; et al. Recombinant Human Collagen for Tissue Engineered Corneal Substitutes. Biomaterials 2008, 29, 1147–1158. [Google Scholar] [CrossRef]
- Mirazul Islam, M.; Cėpla, V.; He, C.; Edin, J.; Rakickas, T.; Kobuch, K.; Ruželė, Ž.; Bruce Jackson, W.; Rafat, M.; Lohmann, C.P.; et al. Functional Fabrication of Recombinant Human Collagen-Phosphorylcholine Hydrogels for Regenerative Medicine Applications. Acta Biomater. 2015, 12, 70–80. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Griffith, M.; Watsky, M.A.; Forrester, J.V.; Kuffová, L.; Grant, D.; Merrett, K.; Carlsson, D.J. Properties of Porcine and Recombinant Human Collagen Matrices for Optically Clear Tissue Engineering Applications. Biomacromolecules 2006, 7, 1819–1828. [Google Scholar] [CrossRef]
- Liu, W.; Deng, C.; McLaughlin, C.R.; Fagerholm, P.; Lagali, N.S.; Heyne, B.; Scaiano, J.C.; Watsky, M.A.; Kato, Y.; Munger, R.; et al. Collagen-Phosphorylcholine Interpenetrating Network Hydrogels as Corneal Substitutes. Biomaterials 2009, 30, 1551–1559. [Google Scholar] [CrossRef]
- Xeroudaki, M.; Thangavelu, M.; Lennikov, A.; Ratnayake, A.; Bisevac, J.; Petrovski, G.; Fagerholm, P.; Rafat, M.; Lagali, N. A Porous Collagen-Based Hydrogel and Implantation Method for Corneal Stromal Regeneration and Sustained Local Drug Delivery. Sci. Rep. 2020, 10, 16936. [Google Scholar] [CrossRef] [PubMed]
- McLaughlin, S.; McNeill, B.; Podrebarac, J.; Hosoyama, K.; Sedlakova, V.; Cron, G.; Smyth, D.; Seymour, R.; Goel, K.; Liang, W.; et al. Injectable Human Recombinant Collagen Matrices Limit Adverse Remodeling and Improve Cardiac Function after Myocardial Infarction. Nat. Commun. 2019, 10, 4866. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Debelle, L.; Tamburro, A.M. Elastin: Molecular Description and Function. Int. J. Biochem. Cell Biol. 1999, 31, 261–272. [Google Scholar] [CrossRef] [PubMed]
- MacEwan, S.R.; Chilkoti, A. Elastin-like Polypeptides: Biomedical Applications of Tunable Biopolymers. Pept. Sci. 2010, 94, 60–77. [Google Scholar] [CrossRef]
- Wang, R.; Yang, Z.; Luo, J.; Hsing, I.-M.; Sun, F. B12-Dependent Photoresponsive Protein Hydrogels for Controlled Stem Cell/Protein Release. Proc. Natl. Acad. Sci. USA 2017, 114, 5912–5917. [Google Scholar] [CrossRef] [Green Version]
- Jiang, B.; Liu, X.; Yang, C.; Yang, Z.; Luo, J.; Kou, S.; Liu, K.; Sun, F. Injectable, Photoresponsive Hydrogels for Delivering Neuroprotective Proteins Enabled by Metal-Directed Protein Assembly. Sci. Adv. 2020, 6, eabc4824. [Google Scholar] [CrossRef]
- Leal-Egaña, A.; Scheibel, T. Silk-Based Materials for Biomedical Applications. Biotechnol. Appl. Biochem. 2010, 55, 155–167. [Google Scholar] [CrossRef] [PubMed]
- Floren, M.; Migliaresi, C.; Motta, A. Processing Techniques and Applications of Silk Hydrogels in Bioengineering. J. Funct. Biomater. 2016, 7, 26. [Google Scholar] [CrossRef] [Green Version]
- Schacht, K.; Jüngst, T.; Schweinlin, M.; Ewald, A.; Groll, J.; Scheibel, T. Biofabrication of Cell-Loaded 3D Spider Silk Constructs. Angew. Chem. Int. Ed. 2015, 54, 2816–2820. [Google Scholar] [CrossRef]
- Koh, K.; Wang, J.K.; Chen, J.X.Y.; Hiew, S.H.; Cheng, H.S.; Gabryelczyk, B.; Vos, M.I.G.; Yip, Y.S.; Chen, L.; Sobota, R.M.; et al. Squid Suckerin-Spider Silk Fusion Protein Hydrogel for Delivery of Mesenchymal Stem Cell Secretome to Chronic Wounds. Adv. Healthc. Mater. 2023, 12, 2201900. [Google Scholar] [CrossRef]
- Hu, X.; Xia, X.-X.; Huang, S.-C.; Qian, Z.-G. Development of Adhesive and Conductive Resilin-Based Hydrogels for Wearable Sensors. Biomacromolecules 2019, 20, 3283–3293. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Tong, Z.; Jia, X.; Kiick, K.L. Resilin-Like Polypeptide Hydrogels Engineered for Versatile Biological Functions. Soft Matter 2013, 9, 665–673. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rouse, J.G.; Van Dyke, M.E. A Review of Keratin-Based Biomaterials for Biomedical Applications. Materials 2010, 3, 999–1014. [Google Scholar] [CrossRef] [Green Version]
- Kan, J.; Li, W.; Qing, R.; Gao, F.; Wang, Y.; Zhu, L.; Wang, B.; Hao, S. Study of Mechanisms of Recombinant Keratin Solubilization with Enhanced Wound Healing Capability. Chem. Mater. 2020, 32, 3122–3133. [Google Scholar] [CrossRef]
- Tomblyn, S.; Pettit Kneller, E.L.; Walker, S.J.; Ellenburg, M.D.; Kowalczewski, C.J.; Van Dyke, M.; Burnett, L.; Saul, J.M. Keratin Hydrogel Carrier System for Simultaneous Delivery of Exogenous Growth Factors and Muscle Progenitor Cells. J. Biomed. Mater. Res. B Appl. Biomater. 2016, 104, 864–879. [Google Scholar] [CrossRef] [Green Version]
- MacEwan, S.R.; Chilkoti, A. Applications of Elastin-like Polypeptides in Drug Delivery. J. Control Release 2014, 190, 314–330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, A.; Li, Y.; Yao, Y.; Yang, X.; Cao, Z.; Nie, H.; Yang, G. Injectable Keratin Hydrogels as Hemostatic and Wound Dressing Materials. Biomater. Sci. 2021, 9, 4169–4177. [Google Scholar] [CrossRef] [PubMed]
- Newman, K.; Clark, K.; Gurumurthy, B.; Pal, P.; Janorkar, A.V. Elastin-Collagen Based Hydrogels as Model Scaffolds to Induce Three-Dimensional Adipocyte Culture from Adipose Derived Stem Cells. Bioengineering 2020, 7, 110. [Google Scholar] [CrossRef]
- Wang, S.; Wang, Z.; Foo, S.E.M.; Tan, N.S.; Yuan, Y.; Lin, W.; Zhang, Z.; Ng, K.W. Culturing Fibroblasts in 3D Human Hair Keratin Hydrogels. ACS Appl. Mater. Interfaces 2015, 7, 5187–5198. [Google Scholar] [CrossRef] [PubMed]
- Developing a Cas9-Based Tool to Engineer Native Plasmids in Synechocystis Sp. PCC 6803—Xiao—2018—Biotechnology and Bioengineering—Wiley Online Library. Available online: https://onlinelibrary.wiley.com/doi/abs/10.1002/bit.26747 (accessed on 24 May 2023).
- Tang, T.-C.; An, B.; Huang, Y.; Vasikaran, S.; Wang, Y.; Jiang, X.; Lu, T.K.; Zhong, C. Materials Design by Synthetic Biology. Nat. Rev. Mater. 2021, 6, 332–350. [Google Scholar] [CrossRef]
- Raftery, E.D.; Gharkhanian, E.G.; Ricapito, N.G.; McNamara, J.; Deming, T.J. Influence of Sulfur-Containing Diamino Acid Structure on Covalently Crosslinked Copolypeptide Hydrogels. Chem. Asian J. 2018, 13, 3547–3553. [Google Scholar] [CrossRef]
- Jervis, P.J.; Amorim, C.; Pereira, T.; Martins, J.A.; Ferreira, P.M.T. Dehydropeptide Supramolecular Hydrogels and Nanostructures as Potential Peptidomimetic Biomedical Materials. Int. J. Mol. Sci. 2021, 22, 2528. [Google Scholar] [CrossRef]
- Bai, W.; Geng, W.; Wang, S.; Zhang, F. Biosynthesis, Regulation, and Engineering of Microbially Produced Branched Biofuels. Biotechnol. Biofuels 2019, 12, 84. [Google Scholar] [CrossRef] [Green Version]
- Jiang, W.; Gu, P.; Zhang, F. Steps towards “drop-in” Biofuels: Focusing on Metabolic Pathways. Curr. Opin. Biotechnol. 2018, 53, 26–32. [Google Scholar] [CrossRef]
- Jiang, W.; Qiao, J.B.; Bentley, G.J.; Liu, D.; Zhang, F. Modular Pathway Engineering for the Microbial Production of Branched-Chain Fatty Alcohols. Biotechnol. Biofuels 2017, 10, 244. [Google Scholar] [CrossRef]
- Bai, W.; Anthony, W.E.; Hartline, C.J.; Wang, S.; Wang, B.; Ning, J.; Hsu, F.-F.; Dantas, G.; Zhang, F. Engineering Diverse Fatty Acid Compositions of Phospholipids in Escherichia Coli. Metab. Eng. 2022, 74, 11–23. [Google Scholar] [CrossRef]
- Schmitz, A.C.; Hartline, C.J.; Zhang, F. Engineering Microbial Metabolite Dynamics and Heterogeneity. Biotechnol. J. 2017, 12, 1700422. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Zhang, F. Metabolic Feedback Circuits Provide Rapid Control of Metabolite Dynamics. ACS Synth. Biol. 2018, 7, 347–356. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Zhang, F. Control Strategies to Manage Trade-Offs during Microbial Production. Curr. Opin. Biotechnol. 2020, 66, 158–164. [Google Scholar] [CrossRef]
- Griffith, J.E.; Chen, Y.; Liu, Q.; Wang, Q.; Richards, J.J.; Tullman-Ercek, D.; Shull, K.R.; Wang, M. Quantitative High-Throughput Measurement of Bulk Mechanical Properties Using Commonly Available Equipment. Mater. Horiz. 2023, 10, 97–106. [Google Scholar] [CrossRef] [PubMed]
- Kang, E.; Choi, Y.Y.; Chae, S.-K.; Moon, J.-H.; Chang, J.-Y.; Lee, S.-H. Microfluidic Spinning of Flat Alginate Fibers with Grooves for Cell-Aligning Scaffolds. Adv. Mater. 2012, 24, 4271–4277. [Google Scholar] [CrossRef] [PubMed]
- Mredha, M.T.I.; Kitamura, N.; Nonoyama, T.; Wada, S.; Goto, K.; Zhang, X.; Nakajima, T.; Kurokawa, T.; Takagi, Y.; Yasuda, K.; et al. Anisotropic Tough Double Network Hydrogel from Fish Collagen and Its Spontaneous in Vivo Bonding to Bone. Biomaterials 2017, 132, 85–95. [Google Scholar] [CrossRef]
- Singh, A.; Orsat, V.; Raghavan, V. Soybean Hydrophobic Protein Response to External Electric Field: A Molecular Modeling Approach. Biomolecules 2013, 3, 168–179. [Google Scholar] [CrossRef] [Green Version]
- Cao, M.; Liao, L.; Zhang, X.; Chen, X.; Peng, S.; Zou, L.; Liang, R.; Liu, W. Electric Field-Driven Fabrication of Anisotropic Hydrogels from Plant Proteins: Microstructure, Gel Performance and Formation Mechanism. Food Hydrocoll. 2023, 136, 108297. [Google Scholar] [CrossRef]
- Lu, Q.; Bai, S.; Ding, Z.; Guo, H.; Shao, Z.; Zhu, H.; Kaplan, D.L. Hydrogel Assembly with Hierarchical Alignment by Balancing Electrostatic Forces. Adv. Mater. Interfaces 2016, 3, 1500687. [Google Scholar] [CrossRef]
- Ding, Z.; Lu, G.; Cheng, W.; Xu, G.; Zuo, B.; Lu, Q.; Kaplan, D.L. Tough Anisotropic Silk Nanofiber Hydrogels with Osteoinductive Capacity. ACS Biomater. Sci. Eng. 2020, 6, 2357–2367. [Google Scholar] [CrossRef] [PubMed]
- Mredha, M.T.I.; Jeon, I. Biomimetic Anisotropic Hydrogels: Advanced Fabrication Strategies, Extraordinary Functionalities, and Broad Applications. Prog. Mater. Sci. 2022, 124, 100870. [Google Scholar] [CrossRef]
- van Oosten, A.S.G.; Chen, X.; Chin, L.; Cruz, K.; Patteson, A.E.; Pogoda, K.; Shenoy, V.B.; Janmey, P.A. Emergence of Tissue-like Mechanics from Fibrous Networks Confined by Close-Packed Cells. Nature 2019, 573, 96–101. [Google Scholar] [CrossRef]
- Huang, P.-S.; Ban, Y.-E.A.; Richter, F.; Andre, I.; Vernon, R.; Schief, W.R.; Baker, D. RosettaRemodel: A Generalized Framework for Flexible Backbone Protein Design. PLoS ONE 2011, 6, e24109. [Google Scholar] [CrossRef] [Green Version]
- Moreira, I.P.; Scott, G.G.; Ulijn, R.V.; Tuttle, T. Computational Prediction of Tripeptide-Dipeptide Co-Assembly. Mol. Phys. 2019, 117, 1151–1163. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Gianti, E.; Percec, S. Machine Learning at the Interface of Polymer Science and Biology: How Far Can We Go? Biomacromolecules 2022, 23, 576–591. [Google Scholar] [CrossRef] [PubMed]
- Anand, N.; Huang, P. Generative Modeling for Protein Structures. In Proceedings of the Advances in Neural Information Processing Systems; Curran Associates, Inc.: Red Hook, NY, USA, 2018; Volume 31. [Google Scholar]
- Eguchi, R.R.; Choe, C.A.; Huang, P.-S. Ig-VAE: Generative Modeling of Protein Structure by Direct 3D Coordinate Generation. PLOS Comput. Biol. 2022, 18, e1010271. [Google Scholar] [CrossRef]
- Anishchenko, I.; Pellock, S.J.; Chidyausiku, T.M.; Ramelot, T.A.; Ovchinnikov, S.; Hao, J.; Bafna, K.; Norn, C.; Kang, A.; Bera, A.K.; et al. De Novo Protein Design by Deep Network Hallucination. Nature 2021, 600, 547–552. [Google Scholar] [CrossRef]
- Norn, C.; Wicky, B.I.M.; Juergens, D.; Liu, S.; Kim, D.; Koepnick, B.; Anishchenko, I.; Players, F.; Baker, D.; Ovchinnikov, S. Protein sequence design by conformational landscape optimization. Biophys. Comput. Biol. 2020, 118, e2017228118. [Google Scholar] [CrossRef] [PubMed]
- Rives, A.; Meier, J.; Sercu, T.; Goyal, S.; Lin, Z.; Liu, J.; Guo, D.; Ott, M.; Zitnick, C.L.; Ma, J.; et al. Biological Structure and Function Emerge from Scaling Unsupervised Learning to 250 Million Protein Sequences. Proc. Natl. Acad. Sci. USA 2021, 118, e2016239118. [Google Scholar] [CrossRef]
- Alley, E.C.; Khimulya, G.; Biswas, S.; AlQuraishi, M.; Church, G.M. Unified Rational Protein Engineering with Sequence-Based Deep Representation Learning. Nat. Methods 2019, 16, 1315–1322. [Google Scholar] [CrossRef]




| Hydrogels | Advantage | Disadvantage | References |
|---|---|---|---|
| pH and ion-induced protein hydrogels |
|
| [4,16,17,18,54,55] |
| Temperature-induced hydrogel formation |
|
| [23,25,26] |
| Protein–protein interaction-based protein hydrogels |
|
| [31] |
| Light-controlled protein hydrogel |
|
| [38,56,57] |
| Chemical crosslinker-based protein hydrogel |
|
| [45,58,59] |
| Enzymatic crosslinked hydrogels |
|
| [49,51,52] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, K.Z.; Jeon, J.; Jiang, B.; Subramani, S.V.; Li, J.; Zhang, F. Protein-Based Hydrogels and Their Biomedical Applications. Molecules 2023, 28, 4988. https://doi.org/10.3390/molecules28134988
Lee KZ, Jeon J, Jiang B, Subramani SV, Li J, Zhang F. Protein-Based Hydrogels and Their Biomedical Applications. Molecules. 2023; 28(13):4988. https://doi.org/10.3390/molecules28134988
Chicago/Turabian StyleLee, Kok Zhi, Juya Jeon, Bojing Jiang, Shri Venkatesh Subramani, Jingyao Li, and Fuzhong Zhang. 2023. "Protein-Based Hydrogels and Their Biomedical Applications" Molecules 28, no. 13: 4988. https://doi.org/10.3390/molecules28134988
APA StyleLee, K. Z., Jeon, J., Jiang, B., Subramani, S. V., Li, J., & Zhang, F. (2023). Protein-Based Hydrogels and Their Biomedical Applications. Molecules, 28(13), 4988. https://doi.org/10.3390/molecules28134988