Phosphoproteome Profiling of uEVs Reveals p-AQP2 and p-GSK3β as Potential Markers for Diabetic Nephropathy
Abstract
:1. Introduction
2. Results
2.1. Clinical Characteristics of the Study Group
2.2. uEVs Isolation and Characterization
2.3. Ultra-High Sensitivity Quantitative Phosphoproteomic Analysis
2.4. Gene Ontology Analysis
2.5. Protein-Protein Interaction Prediction of the Phosphoproteins in uEVs from Diabetic Patients Compared to Patients with DN
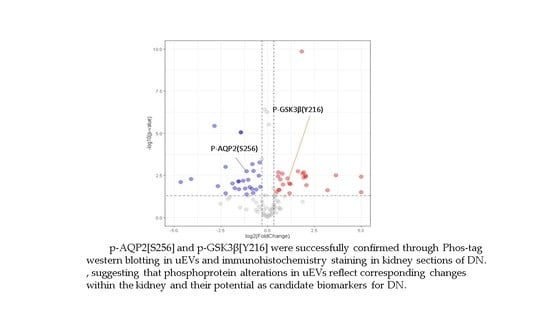
2.6. Validation of Identified Phosphorylation Specific to DN Patients by Phos-Tag Western Blotting and Immunohistochemistry
3. Discussion
4. Materials and Methods
4.1. Patient Recruitment
4.2. Urine Collection and Pre-Processing
4.3. uEVs Separation and Characterization
4.3.1. Urinary Extracellular Vesicle Separation
4.3.2. Transmission Electron Microscopy
4.3.3. Nanoparticle Tracking Analysis
4.3.4. Western Blot Analysis
4.4. Protein Digestion
4.5. Phosphopeptide Enrichment
4.6. Three-Dimensional RP-SAX-RP Systems
4.7. LC-MS/MS Analysis
4.8. Data Processing
4.8.1. Data Processing
4.8.2. Phosphorylation Site Localization
4.8.3. Quantitative Data Analysis
4.9. Immunohistochemistry Stain
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Rayego-Mateos, S.; Rodrigues-Diez, R.R.; Fernandez-Fernandez, B.; Mora-Fernández, C.; Marchant, V.; Donate-Correa, J.; Navarro-González, J.F.; Ortiz, A.; Ruiz-Ortega, M. Targeting inflammation to treat diabetic kidney disease: The road to 2030. Kidney Int. 2023, 103, 282–296. [Google Scholar] [CrossRef] [PubMed]
- Tong, X.; Yu, Q.; Ankawi, G.; Pang, B.; Yang, B.; Yang, H. Insights into the Role of Renal Biopsy in Patients with T2DM: A Literature Review of Global Renal Biopsy Results. Diabetes Ther. Res. Treat. Educ. Diabetes Relat. Disord. 2020, 11, 1983–1999. [Google Scholar] [CrossRef] [PubMed]
- Klessens, C.Q.; Woutman, T.D.; Veraar, K.A.; Zandbergen, M.; Valk, E.J.; Rotmans, J.I.; Wolterbeek, R.; Bruijn, J.A.; Bajema, I.M. An autopsy study suggests that diabetic nephropathy is underdiagnosed. Kidney Int. 2016, 90, 149–156. [Google Scholar] [CrossRef] [PubMed]
- Betz, B.B.; Jenks, S.J.; Cronshaw, A.D.; Lamont, D.J.; Cairns, C.; Manning, J.R.; Goddard, J.; Webb, D.J.; Mullins, J.J.; Hughes, J.; et al. Urinary peptidomics in a rodent model of diabetic nephropathy highlights epidermal growth factor as a biomarker for renal deterioration in patients with type 2 diabetes. Kidney Int. 2016, 89, 1125–1135. [Google Scholar] [CrossRef]
- Persson, F.; Rossing, P. Diagnosis of diabetic kidney disease: State of the art and future perspective. Kidney Int. Suppl. (2011) 2018, 8, 2–7. [Google Scholar] [CrossRef] [Green Version]
- Merchant, M.L.; Rood, I.M.; Deegens, J.K.J.; Klein, J.B. Isolation and characterization of urinary extracellular vesicles: Implications for biomarker discovery. Nat. Rev. Nephrol. 2017, 13, 731–749. [Google Scholar] [CrossRef] [Green Version]
- Delrue, C.; De Bruyne, S.; Speeckaert, R.; Speeckaert, M.M. Urinary Extracellular Vesicles in Chronic Kidney Disease: From Bench to Bedside? Diagnostics 2023, 13, 443. [Google Scholar] [CrossRef]
- Wu, Q.; Poulsen, S.B.; Murali, S.K.; Grimm, P.R.; Su, X.-T.; Delpire, E.; Welling, P.A.; Ellison, D.H.; Fenton, R.A. Large-Scale Proteomic Assessment of Urinary Extracellular Vesicles Highlights Their Reliability in Reflecting Protein Changes in the Kidney. J. Am. Soc. Nephrol. 2021, 32, 2195–2209. [Google Scholar] [CrossRef]
- Gudehithlu, K.P.; Garcia-Gomez, I.; Vernik, J.; Brecklin, C.; Kraus, M.; Cimbaluk, D.J.; Hart, P.; Dunea, G.; Arruda, J.A.; Singh, A.K. In Diabetic Kidney Disease Urinary Exosomes Better Represent Kidney Specific Protein Alterations Than Whole Urine. Am. J. Nephrol. 2015, 42, 418–424. [Google Scholar] [CrossRef]
- Gudehithlu, K.P.; Hart, P.; Joshi, A.; Garcia-Gomez, I.; Cimbaluk, D.J.; Dunea, G.; Arruda, J.A.L.; Singh, A.K. Urine exosomal ceruloplasmin: A potential early biomarker of underlying kidney disease. Clin. Exp. Nephrol. 2019, 23, 1013–1021. [Google Scholar] [CrossRef]
- De, S.; Kuwahara, S.; Hosojima, M.; Ishikawa, T.; Kaseda, R.; Sarkar, P.; Yoshioka, Y.; Kabasawa, H.; Iida, T.; Goto, S.; et al. Exocytosis-Mediated Urinary Full-Length Megalin Excretion Is Linked With the Pathogenesis of Diabetic Nephropathy. Diabetes 2017, 66, 1391–1404. [Google Scholar] [CrossRef] [Green Version]
- Sinha, N.; Kumar, V.; Puri, V.; Nada, R.; Rastogi, A.; Jha, V.; Puri, S. Urinary exosomes: Potential biomarkers for diabetic nephropathy. Nephrology (Carlton, Vic.) 2020, 25, 881–887. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rossi, L.; Nicoletti, M.C.; Carmosino, M.; Mastrofrancesco, L.; Di Franco, A.; Indrio, F.; Lella, R.; Laviola, L.; Giorgino, F.; Svelto, M.; et al. Urinary Excretion of Kidney Aquaporins as Possible Diagnostic Biomarker of Diabetic Nephropathy. J. Diabetes Res. 2017, 2017, 4360357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rachubik, P.; Piwkowska, A. The role of vasodilator-stimulated phosphoprotein in podocyte functioning. Cell Biol. Int. 2019, 43, 1092–1101. [Google Scholar] [CrossRef] [PubMed]
- Tejada, T.; Catanuto, P.; Ijaz, A.; Santos, J.V.; Xia, X.; Sanchez, P.; Sanabria, N.; Lenz, O.; Elliot, S.J.; Fornoni, A. Failure to phosphorylate AKT in podocytes from mice with early diabetic nephropathy promotes cell death. Kidney Int. 2008, 73, 1385–1393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, I.H.; Xue, L.; Hsu, C.C.; Paez, J.S.; Pan, L.; Andaluz, H.; Wendt, M.K.; Iliuk, A.B.; Zhu, J.K.; Tao, W.A. Phosphoproteins in extracellular vesicles as candidate markers for breast cancer. Proc. Natl. Acad. Sci. USA 2017, 114, 3175–3180. [Google Scholar] [CrossRef]
- Ellis, R.; Katerelos, M.; Choy, S.W.; Cook, N.; Lee, M.; Paizis, K.; Pell, G.; Walker, S.; Power, D.A.; Mount, P.F. Increased expression and phosphorylation of 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase isoforms in urinary exosomes in pre-eclampsia. J. Translat. Med. 2019, 17, 60. [Google Scholar] [CrossRef]
- Giudice, G.; Petsalaki, E. Proteomics and phosphoproteomics in precision medicine: Applications and challenges. Brief. Bioinf. 2017, 20, 767–777. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weeraphan, C.; Phongdara, A.; Chaiyawat, P.; Diskul-Na-Ayudthaya, P.; Chokchaichamnankit, D.; Verathamjamras, C.; Netsirisawan, P.; Yingchutrakul, Y.; Roytrakul, S.; Champattanachai, V.; et al. Phosphoproteome Profiling of Isogenic Cancer Cell-Derived Exosome Reveals HSP90 as a Potential Marker for Human Cholangiocarcinoma. Proteomics 2019, 19, e1800159. [Google Scholar] [CrossRef]
- Nunez Lopez, Y.O.; Iliuk, A.; Petrilli, A.M.; Glass, C.; Casu, A.; Pratley, R.E. Proteomics and Phosphoproteomics of Circulating Extracellular Vesicles Provide New Insights into Diabetes Pathobiology. Int. J. Mol. Sci. 2022, 23, 5779. [Google Scholar] [CrossRef]
- Bodenmiller, B.; Malmstrom, J.; Gerrits, B.; Campbell, D.; Lam, H.; Schmidt, A.; Rinner, O.; Mueller, L.N.; Shannon, P.T.; Pedrioli, P.G.; et al. PhosphoPep—A phosphoproteome resource for systems biology research in Drosophila Kc167 cells. Mol. Syst. Biol. 2007, 3, 139. [Google Scholar] [CrossRef] [PubMed]
- Trogden, K.P.; Zhu, X.; Lee, J.S.; Wright, C.V.E.; Gu, G.; Kaverina, I. Regulation of Glucose-Dependent Golgi-Derived Microtubules by cAMP/EPAC2 Promotes Secretory Vesicle Biogenesis in Pancreatic β Cells. Curr. Biol. CB 2019, 29, 2339–2350.e5. [Google Scholar] [CrossRef] [PubMed]
- Blijdorp, C.J.; Tutakhel, O.A.Z.; Hartjes, T.A.; van den Bosch, T.P.P.; van Heugten, M.H.; Rigalli, J.P.; Willemsen, R.; Musterd-Bhaggoe, U.M.; Barros, E.R.; Carles-Fontana, R.; et al. Comparing Approaches to Normalize, Quantify, and Characterize Urinary Extracellular Vesicles. J. Am. Soc. Nephrol. JASN 2021, 32, 1210–1226. [Google Scholar] [CrossRef] [PubMed]
- Cricrì, G.; Bellucci, L.; Montini, G.; Collino, F. Urinary Extracellular Vesicles: Uncovering the Basis of the Pathological Processes in Kidney-Related Diseases. Int. J. Mol. Sci. 2021, 22, 6507. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Liu, K.; Gao, Y. Phosphoproteins with Stability Against All Urinary Phosphatases as Potential Biomarkers in Urine. Protein Pept. Lett. 2015, 22, 795–800. [Google Scholar] [CrossRef]
- Zheng, H.; Guan, S.; Wang, X.; Zhao, J.; Gao, M.; Zhang, X. Deconstruction of Heterogeneity of Size-Dependent Exosome Subpopulations from Human Urine by Profiling N-Glycoproteomics and Phosphoproteomics Simultaneously. Anal. Chem. 2020, 92, 9239–9246. [Google Scholar] [CrossRef]
- Boukouris, S.; Mathivanan, S. Exosomes in bodily fluids are a highly stable resource of disease biomarkers. Proteom. Clin. Appl. 2015, 9, 358–367. [Google Scholar] [CrossRef] [Green Version]
- Erdbrügger, U.; Blijdorp, C.J.; Bijnsdorp, I.V.; Borràs, F.E.; Burger, D.; Bussolati, B.; Byrd, J.B.; Clayton, A.; Dear, J.W.; Falcón-Pérez, J.M.; et al. Urinary extracellular vesicles: A position paper by the Urine Task Force of the International Society for Extracellular Vesicles. J. Extracell. Vesicles 2021, 10, e12093. [Google Scholar] [CrossRef]
- Salih, M.; Demmers, J.A.; Bezstarosti, K.; Leonhard, W.N.; Losekoot, M.; van Kooten, C.; Gansevoort, R.T.; Peters, D.J.; Zietse, R.; Hoorn, E.J. Proteomics of Urinary Vesicles Links Plakins and Complement to Polycystic Kidney Disease. J. Am. Soc. Nephrol. JASN 2016, 27, 3079–3092. [Google Scholar] [CrossRef] [Green Version]
- Erdbrugger, U.; Le, T.H. Extracellular Vesicles in Renal Diseases: More than Novel Biomarkers? J. Am. Soc. Nephrol. JASN 2016, 27, 12–26. [Google Scholar] [CrossRef] [Green Version]
- Salih, M.; Zietse, R.; Hoorn, E.J. Urinary extracellular vesicles and the kidney: Biomarkers and beyond. Am. J. Physiol. Ren. Physiol. 2014, 306, F1251-9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carreras-Planella, L.; Cucchiari, D.; Cañas, L.; Juega, J.; Franquesa, M.; Bonet, J.; Revuelta, I.; Diekmann, F.; Taco, O.; Lauzurica, R.; et al. Urinary vitronectin identifies patients with high levels of fibrosis in kidney grafts. J. Nephrol. 2021, 34, 861–874. [Google Scholar] [CrossRef] [PubMed]
- Braun, F.; Rinschen, M.; Buchner, D.; Bohl, K.; Plagmann, I.; Bachurski, D.; Richard Späth, M.; Antczak, P.; Göbel, H.; Klein, C.; et al. The proteomic landscape of small urinary extracellular vesicles during kidney transplantation. J. Extracell. Vesicles 2020, 10, e12026. [Google Scholar] [CrossRef]
- Ficarro, S.B.; Zhang, Y.; Lu, Y.; Moghimi, A.R.; Askenazi, M.; Hyatt, E.; Smith, E.D.; Boyer, L.; Schlaeger, T.M.; Luckey, C.J.; et al. Improved Electrospray Ionization Efficiency Compensates for Diminished Chromatographic Resolution and Enables Proteomics Analysis of Tyrosine Signaling in Embryonic Stem Cells. Anal. Chem. 2009, 81, 3440–3447. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Lu, Y.; Ficarro, S.B.; Webber, J.T.; Marto, J.A. Nanoflow Low Pressure High Peak Capacity Single Dimension LC-MS/MS Platform for High-Throughput, In-Depth Analysis of Mammalian Proteomes. Anal. Chem. 2012, 84, 5133–5139. [Google Scholar] [CrossRef] [Green Version]
- Zhou, F.; Sikorski, T.W.; Ficarro, S.B.; Webber, J.T.; Marto, J.A. Online nanoflow reversed phase-strong anion exchange-reversed phase liquid chromatography-tandem mass spectrometry platform for efficient and in-depth proteome sequence analysis of complex organisms. Anal. Chem. 2011, 83, 6996–7005. [Google Scholar] [CrossRef] [Green Version]
- Zhou, F.; Cardoza, J.D.; Ficarro, S.B.; Adelmant, G.O.; Lazaro, J.B.; Marto, J.A. Online nanoflow RP-RP-MS reveals dynamics of multicomponent Ku complex in response to DNA damage. J. Proteome Res. 2010, 9, 6242–6255. [Google Scholar] [CrossRef] [Green Version]
- Gonzales, P.A.; Pisitkun, T.; Hoffert, J.D.; Tchapyjnikov, D.; Star, R.A.; Kleta, R.; Wang, N.S.; Knepper, M.A. Large-scale proteomics and phosphoproteomics of urinary exosomes. J. Am. Soc. Nephrol. JASN 2009, 20, 363–379. [Google Scholar] [CrossRef] [Green Version]
- He, J.; Yang, B. Aquaporins in Renal Diseases. Int. J. Mol. Sci. 2019, 20, 366. [Google Scholar] [CrossRef] [Green Version]
- Bradford, D.; Raghuram, V.; Wilson, J.L.; Chou, C.L.; Hoffert, J.D.; Knepper, M.A.; Pisitkun, T. Use of LC-MS/MS and Bayes’ theorem to identify protein kinases that phosphorylate aquaporin-2 at Ser256. Am. J. Physio. Cell Physiol. 2014, 307, C123–C139. [Google Scholar] [CrossRef] [Green Version]
- Xie, L.; Hoffert, J.D.; Chou, C.L.; Yu, M.J.; Pisitkun, T.; Knepper, M.A.; Fenton, R.A. Quantitative analysis of aquaporin-2 phosphorylation. Am. J. Physiol. Ren. Physiol. 2010, 298, F1018-23. [Google Scholar] [CrossRef] [Green Version]
- Christensen, B.M.; Zelenina, M.; Aperia, A.; Nielsen, S. Localization and regulation of PKA-phosphorylated AQP2 in response to V2-receptor agonist/antagonist treatment. Am. J. Physiol. Renal Physiol. 2000, 278, F29–F42. [Google Scholar] [CrossRef] [PubMed]
- Mariappan, M.M.; Prasad, S.; D’Silva, K.; Cedillo, E.; Sataranatarajan, K.; Barnes, J.L.; Choudhury, G.G.; Kasinath, B.S. Activation of glycogen synthase kinase 3beta ameliorates diabetes-induced kidney injury. J. Biol. Chem. 2014, 289, 35363–35375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hurcombe, J.A.; Hartley, P.; Lay, A.C.; Ni, L.; Bedford, J.J.; Leader, J.P.; Singh, S.; Murphy, A.; Scudamore, C.L.; Marquez, E.; et al. Podocyte GSK3 is an evolutionarily conserved critical regulator of kidney function. Nat. Commun. 2019, 10, 403. [Google Scholar] [CrossRef] [PubMed] [Green Version]







| Groups | Age (Years) | HbA1c (%) | ACR (μg/mg) | eGFR (mL/min/ 1.73 m2) | BUN (mmol/L) | Scr (μmol/L) |
|---|---|---|---|---|---|---|
| NC | 50.20 ± 4.53 | NA | NA | 109 ± 9.08 | 5.62 ± 0.31 | 66.8 ± 5.49 |
| DM | 55.80 ± 9.20 | 6.70 ± 0.24 * | 83.68 ± 6.13 * | 113.40 ± 5.43 ** | 6.56 ± 0.40 * | 81.6 ± 9.91 ** |
| DN | 59. 40 ± 7.74 | 9.46 ± 1.75 | 2418.64 ± 1478.57 | 47.80 ± 14.57 | 8.62 ± 1.49 | 151 ± 28.35 |
| (a) | |||
| Gene | Peptide | Phosphor_Site | p-value |
| ACSA | SWSPPPEVSR | S3:Phospho | 0.00240942 |
| AMNLS | RLSLVPK | S3:Phospho | 0.01164248 |
| BAIP2 | AFPAQTASGFK | T6:Phospho | 0.003610772 |
| BAIP2 | LSDSYSNTLPVR | T8:Phospho | 0.00927998 |
| CHSP1 | GNVVPSPLPTR | S6:Phospho | 0.011014319 |
| ES8L1 | SLNSTPPPPPAPAPAPPPALAR | T5:Phospho | 0.031333883 |
| FIBA | PGSTGTWNPGSSER | S11:Phospho | 0.003771194 |
| GSK3B | GEPNVSYICSR | Y7:Phospho | 0.028989048 |
| GPC5B | TAGFPNGSLGK | S8:Phospho | 0.022155551 |
| GPC5C | ATANSQVMGSANSTLR | S13:Phospho | 0.010074399 |
| K1C13 | MIGFPSSAGSVSPR | S12:Phospho | 0.003025495 |
| KIF12 | SPGQVLPPH | S1:Phospho | 1.37 × 10−10 |
| MAP1A | TLPQEPGK | T1:Phospho | 0.00176674 |
| MILK1 | SPVPSPGSSSPQLQVK | S10:Phospho | 0.022496032 |
| MY15B | AVPSPPPPPIVK | S4:Phospho | 0.001997071 |
| NAT8L | MSVDSRFR | S5:Phospho | 0.029409743 |
| NPT2A | LGSPAVSPLPVR | S7:Phospho | 0.002418861 |
| PDZ1I | YSSMAASFR | S7:Phospho | 0.004247949 |
| SHAN2 | RAPSPVVSPTEMNK | S4:Phospho | 0.035395595 |
| VINEX | AIETRLPSPK | T4:Phospho | 0.003764395 |
| VP37D | SAQPAPTSAADPPK | S1:Phospho | 0.023114006 |
| YG015 | IELSYNK | Y5:Phospho | 0.002020309 |
| ZNF25 | SHFIIHQR | S1:Phospho | 0.003312002 |
| (b) | |||
| AQP2 | RQSVELHSPQSLPR | S3:Phospho | 0.001562435 |
| AQP2 | RRQSVELHSPQSLPR | S4:Phospho | 0.001865546 |
| CAN7L | INSAHGSDKSK | S3:Phospho | 0.005547201 |
| CF170 | KWMHAHYSR | S8:Phospho | 8.55 × 10−6 |
| CHM4C | LPNVPSSSLPAQPNR | S8:Phospho | 0.03478666 |
| CI169 | NPYAHISIPR | S7:Phospho | 0.014805139 |
| CK052 | TLKPQPQQLQQNLPK | T1:Phospho | 0.015894205 |
| CP250 | QSESLSELITLR | S2:Phospho | 0.007130084 |
| CSPP1 | QPSPIVPALQNK | S3:Phospho | 0.000997041 |
| EPS8 | APAPAPPGTVTQVDVR | T9:Phospho | 0.001709885 |
| EPS8 | KGPGEGVLTLR | T9:Phospho | 0.003312002 |
| ES8L2 | HSPTSEPTPPGDALPPVSSPHTHR | S19:Phospho | 0.041726109 |
| EZRI | QLLTLSSELSQAR | T4:Phospho | 3.66 × 10−6 |
| GPC5C | VPSEGAYDIILPR | S3:Phospho | 0.00052554 |
| ICK | STPGLIPRPPAAQPVHGR | T2:Phospho | 0.021340709 |
| IF4E2 | TASDQATTARIR | S3:Phospho | 0.019186167 |
| LDHA | TPKIVSGK | T1:Phospho | 0.020570378 |
| LMNA | SGAQASSTPLSPTRITR | S11:Phospho | 0.00927998 |
| MUC1 | DTYHPMSEYPTYHTHGR | S7:Phospho | 0.035395595 |
| RAB10 | FHTITTSYYR | T3:Phospho | 0.006517696 |
| RAI3 | AHAWPSPYKDYEVK | S6:Phospho | 0.01798913 |
| SARG | ANSALTPPKPESGLTLQESNTPGLR | T6:Phospho | 0.019581944 |
| SHS6L | TPNLDWR | T1:Phospho | 0.000684591 |
| TRPV5 | ASLALPTSSLSR | S11:Phospho | 0.00176674 |
| (c) | |||
| Gene | Peptide | Phosphor_Site | p-value |
| ACSA | SWSPPPEVSR | S3:Phospho | 0.00240942 |
| AMNLS | RLSLVPK | S3:Phospho | 0.01164248 |
| BAIP2 | AFPAQTASGFK | T6:Phospho | 0.003610772 |
| BAIP2 | LSDSYSNTLPVR | T8:Phospho | 0.00927998 |
| CHSP1 | GNVVPSPLPTR | S6:Phospho | 0.011014319 |
| ES8L1 | SLNSTPPPPPAPAPAPPPALAR | T5:Phospho | 0.031333883 |
| FIBA | PGSTGTWNPGSSER | S11:Phospho | 0.003771194 |
| GSK3B | GEPNVSYICSR | Y7:Phospho | 0.028989048 |
| GPC5B | TAGFPNGSLGK | S8:Phospho | 0.022155551 |
| GPC5C | ATANSQVMGSANSTLR | S13:Phospho | 0.010074399 |
| K1C13 | MIGFPSSAGSVSPR | S12:Phospho | 0.003025495 |
| KIF12 | SPGQVLPPH | S1:Phospho | 1.37 × 10−10 |
| MAP1A | TLPQEPGK | T1:Phospho | 0.00176674 |
| MILK1 | SPVPSPGSSSPQLQVK | S10:Phospho | 0.022496032 |
| MY15B | AVPSPPPPPIVK | S4:Phospho | 0.001997071 |
| NAT8L | MSVDSRFR | S5:Phospho | 0.029409743 |
| NPT2A | LGSPAVSPLPVR | S7:Phospho | 0.002418861 |
| PDZ1I | YSSMAASFR | S7:Phospho | 0.004247949 |
| SHAN2 | RAPSPVVSPTEMNK | S4:Phospho | 0.035395595 |
| VINEX | AIETRLPSPK | T4:Phospho | 0.003764395 |
| VP37D | SAQPAPTSAADPPK | S1:Phospho | 0.023114006 |
| YG015 | IELSYNK | Y5:Phospho | 0.002020309 |
| ZNF25 | SHFIIHQR | S1:Phospho | 0.003312002 |
| (d) | |||
| AQP2 | RQSVELHSPQSLPR | S3:Phospho | 0.001562435 |
| AQP2 | RRQSVELHSPQSLPR | S4:Phospho | 0.001865546 |
| CAN7L | INSAHGSDKSK | S3:Phospho | 0.005547201 |
| CF170 | KWMHAHYSR | S8:Phospho | 8.55 × 10−6 |
| CHM4C | LPNVPSSSLPAQPNR | S8:Phospho | 0.03478666 |
| CI169 | NPYAHISIPR | S7:Phospho | 0.014805139 |
| CK052 | TLKPQPQQLQQNLPK | T1:Phospho | 0.015894205 |
| CP250 | QSESLSELITLR | S2:Phospho | 0.007130084 |
| CSPP1 | QPSPIVPALQNK | S3:Phospho | 0.000997041 |
| EPS8 | APAPAPPGTVTQVDVR | T9:Phospho | 0.001709885 |
| EPS8 | KGPGEGVLTLR | T9:Phospho | 0.003312002 |
| ES8L2 | HSPTSEPTPPGDALPPVSSPHTHR | S19:Phospho | 0.041726109 |
| EZRI | QLLTLSSELSQAR | T4:Phospho | 3.66 × 10−6 |
| GPC5C | VPSEGAYDIILPR | S3:Phospho | 0.00052554 |
| ICK | STPGLIPRPPAAQPVHGR | T2:Phospho | 0.021340709 |
| IF4E2 | TASDQATTARIR | S3:Phospho | 0.019186167 |
| LDHA | TPKIVSGK | T1:Phospho | 0.020570378 |
| LMNA | SGAQASSTPLSPTRITR | S11:Phospho | 0.00927998 |
| MUC1 | DTYHPMSEYPTYHTHGR | S7:Phospho | 0.035395595 |
| RAB10 | FHTITTSYYR | T3:Phospho | 0.006517696 |
| RAI3 | AHAWPSPYKDYEVK | S6:Phospho | 0.01798913 |
| SARG | ANSALTPPKPESGLTLQESNTPGLR | T6:Phospho | 0.019581944 |
| SHS6L | TPNLDWR | T1:Phospho | 0.000684591 |
| TRPV5 | ASLALPTSSLSR | S11:Phospho | 0.00176674 |
| (e) | |||
| Gene | Peptide | Phosphor_Site | p-value |
| AQP2 | RQSVELHSPQSLPR | S3:Phospho | 0.00351656 |
| AQP2 | RRQSVELHSPQSLPR | S4:Phospho | 0.003246139 |
| CAN7L | INSAHGSDKSK | S3:Phospho | 0.007011764 |
| CF170 | KWMHAHYSR | S8:Phospho | 3.21 × 10−6 |
| CI169 | NPYAHISIPR | S7:Phospho | 0.003453563 |
| CP250 | QSESLSELITLR | S2:Phospho | 0.00612397 |
| CSPP1 | QPSPIVPALQNK | S3:Phospho | 0.004669614 |
| EPS8 | APAPAPPGTVTQVDVR | T9:Phospho | 0.000581478 |
| EPS8 | KGPGEGVLTLR | T9:Phospho | 0.00052707 |
| ES8L1 | AAGEGLLTLR | T8:Phospho | 1.84 × 10−5 |
| ES8L1 | SLNSTPPPPPAPAPAPPPALAR | S4:Phospho | 0.014008085 |
| ES8L2 | HSPTSEPTPPGDALPPVSSPHTHR | S19:Phospho | 0.040008069 |
| EZRI | QLLTLSSELSQAR | T4:Phospho | 8.73 × 10−6 |
| GPC5C | GVGYETILK | T6:Phospho | 1.68 × 10−8 |
| GPC5C | VPSEGAYDIILPR | S3:Phospho | 0.000326668 |
| GSK3B | GEPNVSYICSR | Y7:Phospho | 0.008989048 |
| HSPB1 | GPSWDPFR | S3:Phospho | 0.047463946 |
| ICK | STPGLIPRPPAAQPVHGR | T2:Phospho | 0.006484348 |
| LDHA | TPKIVSGK | T1:Phospho | 0.020666271 |
| MUC1 | DTYHPMSEYPTYHTHGR | T11:Phospho | 0.025694245 |
| RAB10 | FHTITTSYYR | T3:Phospho | 0.005397277 |
| SARG | ANSALTPPKPESGLTLQESNTPGL | RT6:Phospho | 0.041546421 |
| SHS6L | TPNLDWR | T1:Phospho | 0.003375752 |
| TRRAP | LEPAFLSGLR | S7:Phospho | 0.043482561 |
| YG015 | IELSYNK | Y5:Phospho | 0.001729708 |
| (f) | |||
| AMNLS | RLSLVPK | S3:Phospho | 0.014008085 |
| BAIP2 | AFPAQTASGFK | T6:Phospho | 0.00053512 |
| C2D1A | GPASTPTYSPAPTQPAPR | S9:Phospho | 1.85 × 10−11 |
| DYRK2 | KPSAAAPAAYPTGR | S3:Phospho | 0.001809625 |
| G3P | GALQNIIPASTGAAK | S10:Phospho | 0.022036915 |
| GPC5B | TAGFPNGSLGK | S8:Phospho | 0.011034256 |
| IF4E2 | TASDQATTARIR | S3:Phospho | 0.012911956 |
| IST1 | NISSAQIVGPGPKPEASAK | S3:Phospho | 2.28 × 10−5 |
| K1522 | ASPVPAPSSGLHAAVR | S2:Phospho | 0.017621205 |
| KIF12 | DLLSLGSPR | S7:Phospho | 0.000979657 |
| KIF12 | SPGQVLPPH | S1:Phospho | 1.17 × 10−10 |
| LMNA | SGAQASSTPLSPTRITR | S11:Phospho | 0.033756475 |
| MAP1A | TLPQEPGK | T1:Phospho | 0.037174836 |
| MILK1 | SPVPSPGSSSPQLQVK | S10:Phospho | 0.002110621 |
| MY15B | AVPSPPPPPIVK | S4:Phospho | 0.02828527 |
| NPT2A | LGSPAVSPLPVR | S7:Phospho | 0.00017686 |
| SL9A3 | RGSLAFIR | S3:Phospho | 0.031611131 |
| TRPV5 | ASLALPTSSLSR | S11:Phospho | 0.000897584 |
| VP37D | SAQPAPTSAADPPK | S1:Phospho | 0.004954683 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Q.; Zhang, J.; Fang, Y.; Dai, Y.; Jia, P.; Shen, Z.; Xu, S.; Ding, X.; Zhou, F. Phosphoproteome Profiling of uEVs Reveals p-AQP2 and p-GSK3β as Potential Markers for Diabetic Nephropathy. Molecules 2023, 28, 5605. https://doi.org/10.3390/molecules28145605
Li Q, Zhang J, Fang Y, Dai Y, Jia P, Shen Z, Xu S, Ding X, Zhou F. Phosphoproteome Profiling of uEVs Reveals p-AQP2 and p-GSK3β as Potential Markers for Diabetic Nephropathy. Molecules. 2023; 28(14):5605. https://doi.org/10.3390/molecules28145605
Chicago/Turabian StyleLi, Qing, Jiong Zhang, Yi Fang, Yan Dai, Ping Jia, Ziyan Shen, Sujuan Xu, Xiaoqiang Ding, and Feng Zhou. 2023. "Phosphoproteome Profiling of uEVs Reveals p-AQP2 and p-GSK3β as Potential Markers for Diabetic Nephropathy" Molecules 28, no. 14: 5605. https://doi.org/10.3390/molecules28145605
APA StyleLi, Q., Zhang, J., Fang, Y., Dai, Y., Jia, P., Shen, Z., Xu, S., Ding, X., & Zhou, F. (2023). Phosphoproteome Profiling of uEVs Reveals p-AQP2 and p-GSK3β as Potential Markers for Diabetic Nephropathy. Molecules, 28(14), 5605. https://doi.org/10.3390/molecules28145605