Effects of the Coculture Initiation Method on the Production of Secondary Metabolites in Bioreactor Cocultures of Penicillium rubens and Streptomyces rimosus
Abstract
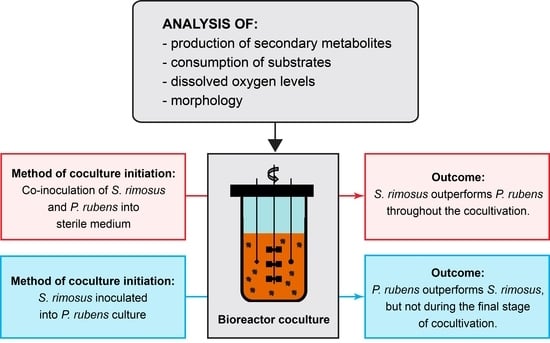
:1. Introduction
2. Results
2.1. Secondary Metabolic Repertoire in Mono- and Cocultures
2.2. Production of Secondary Metabolites by S. rimosus
2.3. Production of Secondary Metabolites by P. rubens
2.4. Dissolved Oxygen Profiles
2.5. Kinetics of Substrate Utilization
2.6. Morphological Analysis
3. Discussion
4. Materials and Methods
4.1. Strains
4.2. Bioreactor Cultivation Runs
4.3. Coculture Initiation
4.4. Medium Composition
4.5. Analytical Methods
4.6. Morphological Analysis
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Del Carratore, F.; Hanko, E.K.; Breitling, R.; Takano, E. Biotechnological application of Streptomyces for the production of clinical drugs and other bioactive molecules. Curr. Opin. Biotechnol. 2022, 77, 102762. [Google Scholar] [CrossRef]
- Seca, A.M.L.; Pinto, D.C.G.A. Biological Potential and Medical Use of Secondary Metabolites. Medicines 2019, 6, 66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McClure, R.; Farris, Y.; Danczak, R.; Nelson, W.; Song, H.-S.; Kessell, A.; Lee, J.-Y.; Couvillion, S.; Henry, C.; Jansson, J.K.; et al. Interaction Networks Are Driven by Community-Responsive Phenotypes in a Chitin-Degrading Consortium of Soil Microbes. Msystems 2022, 7, e0037222. [Google Scholar] [CrossRef] [PubMed]
- Collinge, D.B.; Jensen, B.; Jørgensen, H.J. Fungal endophytes in plants and their relationship to plant disease. Curr. Opin. Microbiol. 2022, 69, 102177. [Google Scholar] [CrossRef]
- Beinart, R.A. The Significance of Microbial Symbionts in Ecosystem Processes. Msystems 2019, 4, e00127-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Demain, A.L.; Fang, A. The natural functions of secondary metabolites. Adv. Biochem. Eng. Biotechnol. 2000, 69, 1–39. [Google Scholar] [CrossRef]
- Bode, H.B.; Bethe, B.; Höfs, R.; Zeeck, A. Big effects from small changes: Possible ways to explore nature’s chemical diversity. ChemBioChem 2002, 3, 619–627. [Google Scholar] [CrossRef]
- Kim, J.H.; Lee, N.; Hwang, S.; Kim, W.; Lee, Y.; Cho, S.; Palsson, B.O.; Cho, B.K. Discovery of novel secondary metabolites encoded in actinomycete genomes through coculture. J. Ind. Microbiol. Biotechnol. 2021, 48, 1. [Google Scholar] [CrossRef]
- Wang, G.; Ran, H.; Fan, J.; Keller, N.P.; Liu, Z.; Wu, F.; Yin, W.B. Fungal-fungal cocultivation leads to widespread secondary metabolite alteration requiring the partial loss-of-function VeA1 protein. Sci. Adv. 2022, 8, 6094. [Google Scholar] [CrossRef]
- Arora, D.; Gupta, P.; Jaglan, S.; Roullier, C.; Grovel, O.; Bertrand, S. Expanding the chemical diversity through microorganisms co-culture: Current status and outlook. Biotechnol. Adv. 2020, 40, 107521. [Google Scholar] [CrossRef]
- Peng, X.Y.; Wu, J.T.; Shao, C.L.; Li, Z.Y.; Chen, M.; Wang, C.Y. Co-culture: Stimulate the metabolic potential and explore the molecular diversity of natural products from microorganisms. Mar. Life Sci. Technol. 2021, 3, 363–374. [Google Scholar] [CrossRef] [PubMed]
- Sonnenbichler, J.; Bliestle, I.M.; Peipp, H.; Holdenrieder, O. Secondary fungal metabolites and their biological activities, i. isolation of antibiotic compounds from cultures of Heterobasidion annosum synthesized in the presence of antagonistic fungi or host plant cells. Biol. Chem. Hoppe Seyler 1989, 370, 1295–1304. [Google Scholar] [CrossRef]
- Jomori, T.; Hara, Y.; Sasaoka, M.; Harada, K.; Setiawan, A.; Hirata, K.; Kimishima, A.; Arai, M. Mycobacterium smegmatis alters the production of secondary metabolites by marine-derived Aspergillus niger. J. Nat. Med. 2020, 74, 76–82. [Google Scholar] [CrossRef] [PubMed]
- Luti, K.J.K.; Mavituna, F. Elicitation of Streptomyces coelicolor with E. coli in a bioreactor enhances undecylprodigiosin production. Biochem. Eng. J. 2011, 53, 281–285. [Google Scholar] [CrossRef]
- Fierro, F.; Vaca, I.; Castillo, N.I.; García-Rico, R.O.; Chávez, R. Penicillium chrysogenum, a Vintage Model with a Cutting-Edge Profile in Biotechnology. Microorganisms 2022, 10, 573. [Google Scholar] [CrossRef] [PubMed]
- Boruta, T.; Milczarek, I.; Bizukojc, M. Evaluating the outcomes of submerged co-cultivation: Production of lovastatin and other secondary metabolites by Aspergillus terreus in fungal co-cultures. Appl. Microbiol. Biotechnol. 2019, 103, 5593–5605. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boruta, T.; Marczyk, A.; Rychta, K.; Przydacz, K.; Bizukojc, M. Confrontation between Penicillium rubens and Aspergillus terreus: Investigating the production of fungal secondary metabolites in submerged co-cultures. J. Biosci. Bioeng. 2020, 130, 503–513. [Google Scholar] [CrossRef]
- Petkovic, H.; Lukežic, T.; Šuškovic, J. Biosynthesis of oxytetracycline by Streptomyces rimosus: Past, present and future directions in the development of tetracycline antibiotics. Food Technol. Biotechnol. 2017, 55, 3–13. [Google Scholar] [CrossRef]
- Boruta, T.; Ścigaczewska, A. Enhanced oxytetracycline production by Streptomyces rimosus in submerged co-cultures with Streptomyces noursei. Molecules 2021, 26, 6036. [Google Scholar] [CrossRef]
- Boruta, T.; Ścigaczewska, A.; Bizukojć, M. “Microbial Wars” in a Stirred Tank Bioreactor: Investigating the Co-Cultures of Streptomyces rimosus and Aspergillus terreus, Filamentous Microorganisms Equipped With a Rich Arsenal of Secondary Metabolites. Front. Bioeng. Biotechnol. 2021, 9, 713639. [Google Scholar] [CrossRef]
- Zygmunt, W.A. Oxytetracycline Formation by Streptomyces rimosus in Chemically Defined Media. Appl. Microbiol. 1961, 9, 502–507. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Song, Z.; Ma, Z.; Bechthold, A.; Yu, X. Sequential improvement of rimocidin production in Streptomyces rimosus M527 by introduction of cumulative drug-resistance mutations. J. Ind. Microbiol. Biotechnol. 2019, 46, 697–708. [Google Scholar] [CrossRef]
- Seco, E.M.; Pérez-Zúñiga, F.J.; Rolón, M.S.; Malpartida, F. Starter unit choice determines the production of two tetraene macrolides, rimocidin and CE-108, in Streptomyces diastaticus var. 108. Chem. Biol. 2004, 11, 357–366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seco, E.M.; Cuesta, T.; Fotso, S.; Laatsch, H.; Malpartida, F. Two polyene amides produced by genetically modified Streptomyces diastaticus var. 108. Chem. Biol. 2005, 12, 535–543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takiguchi, Y.; Terao, M.; Mishima, H.; Okuda, M.; Aoki, A.; Fukuda, R. Milbemycins, a new family of macrolide antibiotics: Fermentation, isolation and physico-chemical properties. J. Antibiot. 1980, 33, 1120–1127. [Google Scholar] [CrossRef] [Green Version]
- Kodani, S.; Bicz, J.; Song, L.; Deeth, R.J.; Ohnishi-Kameyama, M.; Yoshida, M.; Ochi, K.; Challis, G.L. Structure and biosynthesis of scabichelin, a novel tris-hydroxamate siderophore produced by the plant pathogen Streptomyces scabies 87.22. Org. Biomol. Chem. 2013, 11, 4686–4694. [Google Scholar] [CrossRef] [Green Version]
- Armstrong, D.J.; Scarbrough, E.; Skoog, F.; Cole, D.L.; Leonard, N.J. Cytokinins in Corynebacterium fascians Cultures. Plant Physiol. 1976, 58, 749–752. [Google Scholar] [CrossRef] [Green Version]
- McClure, R.A.; Goering, A.W.; Ju, K.S.; Baccile, J.A.; Schroeder, F.C.; Metcalf, W.W.; Thomson, R.J.; Kelleher, N.L. Elucidating the Rimosamide-Detoxin Natural Product Families and Their Biosynthesis Using Metabolite/Gene Cluster Correlations. ACS Chem. Biol. 2016, 11, 11–3452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manwaring, D.G.; Rickards, R.W.; Gaudiano, G.; Nicolella, V. The biosynthesis of the macrolide antibiotic lucensomycin. J. Antibiot. 1969, 22, 545–550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, P.; La, S.; MacMillan, J.B. 1,3-Oxazin-6-one Derivatives and Bohemamine-Type Pyrrolizidine Alkaloids from a Marine-Derived Streptomyces spinoverrucosus. J. Nat. Prod. 2016, 79, 455–462. [Google Scholar] [CrossRef]
- Wu, C.; van der Heul, H.U.; Melnik, A.V.; Lübben, J.; Dorrestein, P.C.; Minnaard, A.J.; Choi, Y.H.; van Wezel, G.P. Lugdunomycin, an Angucycline-Derived Molecule with Unprecedented Chemical Architecture. Angew. Chemie Int. Ed. 2019, 58, 2809–2814. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hochstein, F.A.; von Wittenau, M.S.; Tanner, F.W.; Murai, K. 2-Acetyl-2-decarboxamidoöxytetracycline. J. Am. Chem. Soc. 1960, 82, 5934–5937. [Google Scholar] [CrossRef]
- Shang, N.N.; Zhang, Z.; Huang, J.P.; Wang, L.; Luo, J.; Yang, J.; Peng, T.; Yan, Y.; Ma, Y.T.; Huang, S.X. Glycosylated piericidins from an endophytic Streptomyces with cytotoxicity and antimicrobial activity. J. Antibiot. 2018, 71, 672–676. [Google Scholar] [CrossRef]
- Houbraken, J.; Frisvad, J.C.; Samson, R.A. Fleming’s penicillin producing strain is not Penicillium chrysogenum but P. rubens. IMA Fungus. 2011, 2, 87–95. [Google Scholar] [CrossRef] [Green Version]
- Seifar, R.M.; Zhao, Z.; van Dam, J.; van Winden, W.; van Gulik, W.; Heijnen, J.J. Quantitative analysis of metabolites in complex biological samples using ion-pair reversed-phase liquid chromatography-isotope dilution tandem mass spectrometry. J. Chromatogr. A 2008, 1187, 103–110. [Google Scholar] [CrossRef] [PubMed]
- Viggiano, A.; Salo, O.; Ali, H.; Szymanski, W.; Lankhorst, P.P.; Nygård, Y.; Bovenberg, R.A.L.; Driessen, A.J.M. Pathway for the biosynthesis of the pigment chrysogine by Penicillium chrysogenum. Appl. Environ. Microbiol. 2018, 84, e02246-17. [Google Scholar] [CrossRef] [Green Version]
- Masaaki, T.; Takeshi, K.; Tsuyoshi, H.; Shuji, T. Adenophostins A And B: Potent Agonists of Inositol- 1,4,5-Trisphosphate Receptor Producedby Penicillium brevicompactum Taxonomy, Fermentation, Isolation, Physico-Chemical and Biological Properties. J. Antibiot. 1993, 46, 1643–1647. [Google Scholar] [CrossRef] [Green Version]
- Xu, X.; Zhang, X.; Nong, X.; Wei, X.; Qi, S. Oxindole alkaloids from the fungus Penicillium commune DFFSCS026 isolated from deep-sea-derived sediments. Tetrahedron 2015, 71, 610–615. [Google Scholar] [CrossRef]
- Duan, R.; Zhou, H.; Yang, Y.; Li, H.; Dong, J.; Li, X.; Chen, G.; Zhao, L.; Ding, Z. Antimicrobial meroterpenoids from the endophytic fungus Penicillium sp. T2-8 associated with Gastrodia elata. Phytochem. Lett. 2016, 18, 197–201. [Google Scholar] [CrossRef]
- Li, Y.; Lee, S.R.; Han, E.J.; Seyedsayamdost, M.R. Momomycin, an Antiproliferative Cryptic Metabolite from the Oxytetracycline Producer Streptomyces rimosus. Angew. Chem. Int. Ed. Engl. 2022, 61, e202208573. [Google Scholar] [CrossRef]
- Kowalska, A.; Boruta, T.; Bizukojć, M. Morphological evolution of various fungal species in the presence and absence of aluminum oxide microparticles: Comparative and quantitative insights into microparticle-enhanced cultivation (MPEC). Microbiologyopen 2018, 7, e00603. [Google Scholar] [CrossRef]
- Wucherpfennig, T.; Hestler, T.; Krull, R. Morphology engineering—Osmolality and its effect on Aspergillus niger morphology and productivity. Microb. Cell Fact. 2011, 10, 58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palacio-Barrera, A.M.; Schlembach, I.; Finger, M.; Büchs, J.; Rosenbaum, M.A. Reliable online measurement of population dynamics for filamentous co-cultures. Microb. Biotechnol. 2022, 15, 2773–2785. [Google Scholar] [CrossRef] [PubMed]
- Schlembach, I.; Grünberger, A.; Rosenbaum, M.A.; Regestein, L. Measurement Techniques to Resolve and Control Population Dynamics of Mixed-Culture Processes. Trends Biotechnol. 2021, 39, 1093–1109. [Google Scholar] [CrossRef] [PubMed]
- Finger, M.; Palacio-Barrera, A.M.; Richter, P.; Schlembach, I.; Rosenbaum, M.A. Tunable population dynamics in a synthetic filamentous co-culture. Microbiologyopen 2022, 11, e1324. [Google Scholar] [CrossRef]
- Boruta, T.; Ścigaczewska, A.; Bizukojć, M. Production of secondary metabolites in stirred tank bioreactor co-cultures of Streptomyces noursei and Aspergillus terreus. Front. Bioeng. Biotechnol. 2022, 10, 1011220. [Google Scholar] [CrossRef]
- Brakhage, A.A. Regulation of fungal secondary metabolism. Nat. Rev. Microbiol. 2013, 11, 21–32. [Google Scholar] [CrossRef]
- Liu, C.; Kakeya, H. Cryptic Chemical Communication: Secondary Metabolic Responses Revealed by Microbial Co-culture. Chem. Asian J. 2020, 15, 327–337. [Google Scholar] [CrossRef]
- Čihák, M.; Kameník, Z.; Šmídová, K.; Bergman, N.; Benada, O.; Kofronová, O.; Petrícková, K.; Bobek, J. Secondary metabolites produced during the germination of Streptomyces coelicolor. Front. Microbiol. 2017, 8, 2495. [Google Scholar] [CrossRef] [Green Version]
- Ścigaczewska, A.; Boruta, T.; Bizukojć, M. Quantitative Morphological Analysis of Filamentous Microorganisms in Cocultures and Monocultures: Aspergillus terreus and Streptomyces rimosus Warfare in Bioreactors. Biomolecules 2021, 11, 1740. [Google Scholar] [CrossRef]
- Bertrand, S.; Azzollini, A.; Schumpp, O.; Bohni, N.; Schrenzel, J.; Monod, M.; Gindro, K.; Wolfender, J.L. Multi-well fungal co-culture for de novo metabolite-induction in time-series studies based on untargeted metabolomics. Mol. Biosyst. 2014, 10, 2289–2298. [Google Scholar] [CrossRef]
- Choi, J.N.; Kim, J.; Lee, M.Y.; Park, D.K.; Hong, Y.S.; Lee, C.H. Metabolomics revealed novel isoflavones and optimal cultivation time of Cordyceps militaris fermentation. J. Agric. Food Chem. 2010, 58, 4258–4267. [Google Scholar] [CrossRef] [PubMed]
- Geurts, B.P.; Neerincx, A.H.; Bertrand, S.; Leemans, M.A.A.P.; Postma, G.J.; Wolfender, J.L.; Cristescu, S.M.; Buydens, L.M.C.; Jansen, J.J. Combining ANOVA-PCA with POCHEMON to analyse micro-organism development in a polymicrobial environment. Anal. Chim. Acta 2017, 963, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Azzollini, A.; Boggia, L.; Boccard, J.; Sgorbini, B.; Lecoultre, N.; Allard, P.M.; Rubiolo, P.; Rudaz, S.; Gindro, K.; Bicchi, C.; et al. Dynamics of metabolite induction in fungal co-cultures by metabolomics at both volatile and non-volatile levels. Front. Microbiol. 2018, 9, 326597. [Google Scholar] [CrossRef] [Green Version]
- Fajardo-Hernández, C.A.; Khan, F.S.T.; Flores-Bocanegra, L.; Prieto-Davó, A.; Wan, B.; Ma, R.; Qader, M.; Villanueva-Silva, R.; Martínez-Cárdenas, A.; López-Lobato, M.A.; et al. Insights into the Chemical Diversity of Selected Fungi from the Tza Itzá Cenote of the Yucatan Peninsula. ACS Omega 2022, 7, 12171–12185. [Google Scholar] [CrossRef]
- Berry, O.; Briand, E.; Bagot, A.; Chaigné, M.; Meslet-Cladière, L.; Wang, J.; Grovel, O.; Jansen, J.J.; Ruiz, N.; Pont, T.R.D.; et al. Deciphering interactions between the marine dinoflagellate Prorocentrum lima and the fungus Aspergillus pseudoglaucus. Environ. Microbiol. 2023, 25, 250–267. [Google Scholar] [CrossRef]
- da Costa Stuart, A.K.; Furuie, J.L.; Cataldi, T.R.; Stuart, R.M.; Zawadneak, M.A.C.; Labate, C.A.; Pimentel, I.C. Fungal consortium of two Beauveria bassiana strains increases their virulence, growth, and resistance to stress: A metabolomic approach. PLoS ONE 2022, 17, e0271460. [Google Scholar] [CrossRef]
- Van Santen, J.A.; Poynton, E.F.; Iskakova, D.; Mcmann, E.; Alsup, T.A.; Clark, T.N.; Fergusson, C.H.; Fewer, D.P.; Hughes, A.H.; Mccadden, C.A.; et al. The Natural Products Atlas 2.0: A database of microbially-derived natural products. Nucleic Acids Res. 2022, 50, D1317–D1323. [Google Scholar] [CrossRef] [PubMed]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.M.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed minimum reporting standards for chemical analysis: Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef] [Green Version]






| Ionization | (m/z)experimental | Retention Time (min) | Suggested Secondary Metabolite | Formula of Suggested Metabolite (Ionized) | (m/z)theoretical | Absolute Error Δ(m/z) | Microbial Source | Level of Metabolite Identification |
|---|---|---|---|---|---|---|---|---|
| ESI− | 459.1427 | 4.33 | Oxytetracycline [21] | C22H23N2O9 | 459.1404 | +0.0023 | S. rimosus | identified |
| ESI− | 766.3990 | 5.91 | Rimocidin [22] | C39H60NO14 | 766.4014 | −0.0024 | S. rimosus | putatively annotated |
| ESI− | 738.3635 | 5.46 | CE-108 [23] | C37H56NO14 | 738.3701 | −0.0066 | S. rimosus | putatively annotated |
| ESI− | 765.4189 | 5.78 | Rimocidin B [24] | C39H61N2O13 | 765.4174 | +0.0015 | S. rimosus | putatively annotated |
| ESI− | 752.3879 | 5.68 | Rimocidin (27-ethyl) [20] | C38H58NO14 | 752.3857 | +0.0022 | S. rimosus | putatively annotated |
| ESI− | 527.2993 | 5.70 | Milbemycin A3 [25] | C31H43O7 | 527.3009 | −0.0016 | S. rimosus | putatively annotated |
| ESI− | 591.2823 | 6.15 | Milbemycin A3 + [4O] [20] | C31H43O11 | 591.2805 | +0.0018 | S. rimosus | putatively annotated |
| ESI− | 593.3038 | 6.67 | Milbemycin β11 + [4O] [20] | C31H45O11 | 593.2962 | +0.0076 | S. rimosus | putatively annotated |
| ESI− | 619.3103 | 6.91 | Turgichelin [26] | C24H43N8O11 | 619.3051 | −0.0052 | S. rimosus | putatively annotated |
| ESI− | 264.0859 | 4.95 | 2-Methylthio-cis-zeatin [27] | C11H14N5OS | 264.0919 | −0.0060 | S. rimosus | putatively annotated |
| ESI− | 603.3022 | 5.29 | Rimosamide A [28] | C30H43N4O9 | 603.3030 | −0.0008 | S. rimosus | putatively annotated |
| ESI+ | 708.3666 | 5.23 | Lucensomycin [29] | C36H54NO13 | 708.3595 | +0.0071 | S. rimosus | putatively annotated |
| ESI− | 263.1064 | 4.68 | Spinoxazine A [30] | C13H15N2O4 | 263.1032 | +0.0032 | S. rimosus | putatively annotated |
| ESI− | 384.1161 | 4.95 | Unnamed angucycline metabolite previously isolated from Streptomyces sp. QL37 [31] | C20H18O7N | 384.1083 | +0.0078 | S. rimosus | putatively annotated |
| ESI− | 458.1409 | 4.68 | 2-Acetyl-2-decarboxamido-oxytetracycline (ADOTC) [32] | C23H24NO9 | 458.1451 | −0.0042 | S. rimosus | putatively annotated |
| ESI− | 724.3525 | 5.24 | 7-Demethyl-glucopiericidin A [33] | C36H54NO14 | 724.3544 | -0.0019 | S. rimosus | putatively annotated |
| ESI− | 333.0928 | 6.47 | Penicillin G [34] | C16H17N2O4S | 333.0909 | +0.0019 | P. rubens | identified |
| ESI− | 351.0989 | 5.72 | Benzylpenicilloic acid [35] | C16H19N2O5S | 351.1015 | -0.0026 | P. rubens | putatively annotated |
| ESI− | 189.0756 | 4.72 | Chrysogine [36] | C10H9N2O2 | 189.0665 | +0.0091 | P. rubens | putatively annotated |
| ESI− | 710.0464 | 4.86 | Adenophostin B [37] | C18H27N5O19P3 | 710.0513 | −0.0049 | P. rubens | putatively annotated |
| ESI− | 335.1042 | 6.48 | Cyclopiamide D [38] | C19H15N2O4 | 335.1031 | +0.0011 | P. rubens | putatively annotated |
| ESI− | 491.2644 | 8.30 | Preaustinoid D [39] | C27H39O8 | 491.2645 | −0.0001 | P. rubens | putatively annotated |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Boruta, T.; Ścigaczewska, A.; Ruda, A.; Bizukojć, M. Effects of the Coculture Initiation Method on the Production of Secondary Metabolites in Bioreactor Cocultures of Penicillium rubens and Streptomyces rimosus. Molecules 2023, 28, 6044. https://doi.org/10.3390/molecules28166044
Boruta T, Ścigaczewska A, Ruda A, Bizukojć M. Effects of the Coculture Initiation Method on the Production of Secondary Metabolites in Bioreactor Cocultures of Penicillium rubens and Streptomyces rimosus. Molecules. 2023; 28(16):6044. https://doi.org/10.3390/molecules28166044
Chicago/Turabian StyleBoruta, Tomasz, Anna Ścigaczewska, Agnieszka Ruda, and Marcin Bizukojć. 2023. "Effects of the Coculture Initiation Method on the Production of Secondary Metabolites in Bioreactor Cocultures of Penicillium rubens and Streptomyces rimosus" Molecules 28, no. 16: 6044. https://doi.org/10.3390/molecules28166044
APA StyleBoruta, T., Ścigaczewska, A., Ruda, A., & Bizukojć, M. (2023). Effects of the Coculture Initiation Method on the Production of Secondary Metabolites in Bioreactor Cocultures of Penicillium rubens and Streptomyces rimosus. Molecules, 28(16), 6044. https://doi.org/10.3390/molecules28166044